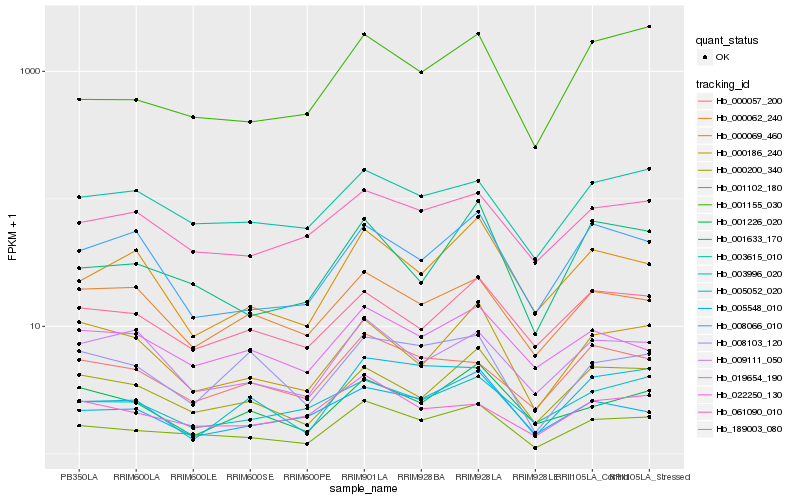
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_180 |
0.0 |
- |
- |
eukaryotic translation initiation factor, putative [Ricinus communis] |
| 2 |
Hb_000200_340 |
0.1259146763 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 3 |
Hb_001633_170 |
0.1296108087 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001155_030 |
0.130928935 |
- |
- |
60S ribosomal protein L10A [Hevea brasiliensis] |
| 5 |
Hb_003615_010 |
0.1342799606 |
- |
- |
60S ribosomal protein L8, putative [Ricinus communis] |
| 6 |
Hb_061090_010 |
0.1357860073 |
- |
- |
RAN BINDING protein 1 [Populus trichocarpa] |
| 7 |
Hb_000057_200 |
0.1388077598 |
- |
- |
PREDICTED: methyltransferase-like protein 22 [Jatropha curcas] |
| 8 |
Hb_000069_460 |
0.1395729787 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 9 |
Hb_000186_240 |
0.1407543973 |
- |
- |
PREDICTED: 39S ribosomal protein L4, mitochondrial [Jatropha curcas] |
| 10 |
Hb_019654_190 |
0.1419797686 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_022250_130 |
0.1423109288 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 12 |
Hb_003996_020 |
0.1430670875 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005548_010 |
0.1432128126 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 14 |
Hb_189003_080 |
0.14491802 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 15 |
Hb_009111_050 |
0.1455539278 |
transcription factor |
TF Family: Jumonji |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540 [Populus euphratica] |
| 16 |
Hb_000062_240 |
0.1456948103 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 17 |
Hb_005052_020 |
0.1460504046 |
- |
- |
- |
| 18 |
Hb_008103_120 |
0.1469117694 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 19 |
Hb_001226_020 |
0.1476108953 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_008066_010 |
0.1477078903 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |