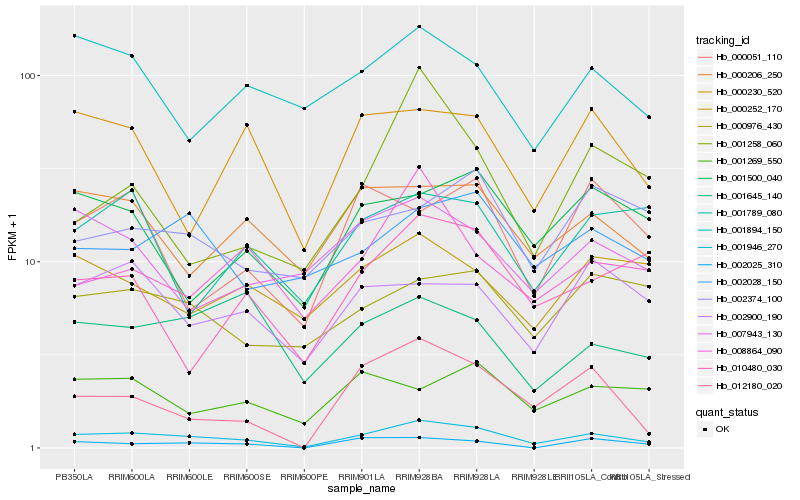
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_270 |
0.0 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 2 |
Hb_012180_020 |
0.1721448913 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |
| 3 |
Hb_000206_250 |
0.182429737 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 4 |
Hb_000230_520 |
0.1877039063 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 5 |
Hb_001894_150 |
0.1917121524 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 6 |
Hb_002028_150 |
0.1922561767 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 7 |
Hb_002900_190 |
0.1929429215 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GTL1 [Jatropha curcas] |
| 8 |
Hb_000976_430 |
0.1941207372 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJD5 [Jatropha curcas] |
| 9 |
Hb_008864_090 |
0.1960366454 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 10 |
Hb_002025_310 |
0.1970457736 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_001500_040 |
0.1972508839 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_007943_130 |
0.1991357 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 13 |
Hb_000252_170 |
0.1997363038 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001645_140 |
0.2010332012 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 15 |
Hb_010480_030 |
0.2020653156 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_002374_100 |
0.2061836341 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 17 |
Hb_001789_080 |
0.2062441582 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 18 |
Hb_000051_110 |
0.206279399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001269_550 |
0.2072195431 |
- |
- |
hypothetical protein POPTR_0001s03510g [Populus trichocarpa] |
| 20 |
Hb_001258_060 |
0.207909756 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |