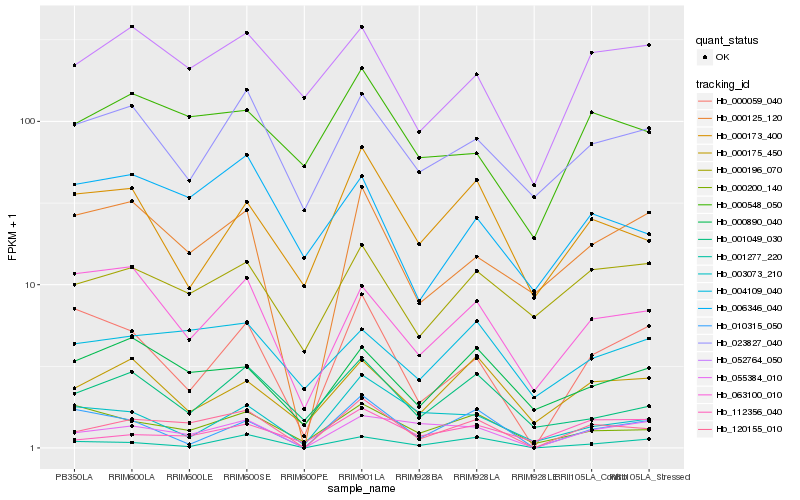
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120155_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_055384_010 |
0.1911613463 |
- |
- |
- |
| 3 |
Hb_000125_120 |
0.2115675342 |
transcription factor |
TF Family: mTERF |
- |
| 4 |
Hb_001277_220 |
0.2160533461 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 5 |
Hb_000059_040 |
0.2171825455 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 6 |
Hb_000200_140 |
0.2237800656 |
- |
- |
PREDICTED: uncharacterized protein LOC104591989 [Nelumbo nucifera] |
| 7 |
Hb_000548_050 |
0.2270149571 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 8 |
Hb_001049_030 |
0.2291426492 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_000890_040 |
0.2314756161 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g15720 [Jatropha curcas] |
| 10 |
Hb_052764_050 |
0.2334924272 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 11 |
Hb_063100_010 |
0.233709804 |
- |
- |
TATA box-binding protein-associated factor [Populus trichocarpa] |
| 12 |
Hb_000196_070 |
0.236077613 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH80-like [Jatropha curcas] |
| 13 |
Hb_004109_040 |
0.2373645066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000175_450 |
0.2375104739 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g52620 [Jatropha curcas] |
| 15 |
Hb_023827_040 |
0.2381497426 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 16 |
Hb_112356_040 |
0.2387207787 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 17 |
Hb_006346_040 |
0.2399757498 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 18 |
Hb_003073_210 |
0.2402724182 |
- |
- |
hypothetical protein CISIN_1g036722mg, partial [Citrus sinensis] |
| 19 |
Hb_010315_050 |
0.2411451267 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 20 |
Hb_000173_400 |
0.2419605008 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |