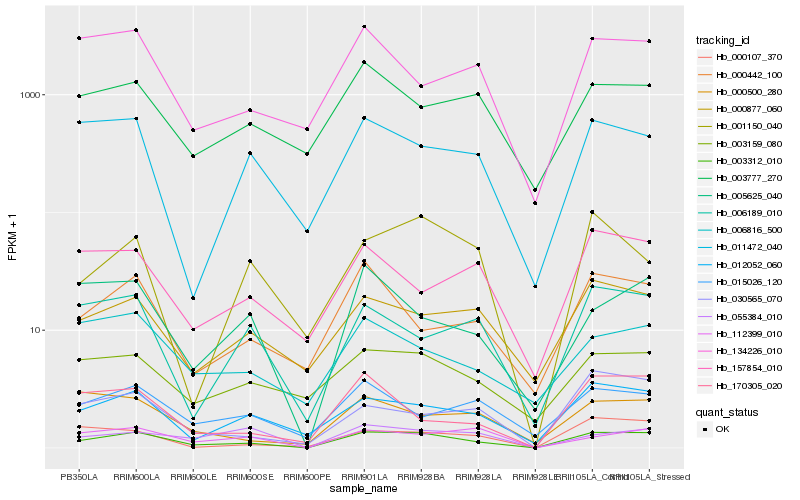
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003312_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |
| 2 |
Hb_012052_060 |
0.1649695151 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 3 |
Hb_000877_060 |
0.1969113063 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 4 |
Hb_003159_080 |
0.1973944351 |
- |
- |
PREDICTED: uncharacterized protein LOC105639276 [Jatropha curcas] |
| 5 |
Hb_005625_040 |
0.1986507211 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 6 |
Hb_000442_100 |
0.1997171848 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 7 |
Hb_112399_010 |
0.2036798345 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_030565_070 |
0.204076652 |
- |
- |
- |
| 9 |
Hb_006189_010 |
0.2041364976 |
- |
- |
- |
| 10 |
Hb_170305_020 |
0.2042342777 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 11 |
Hb_011472_040 |
0.2053023384 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 12 |
Hb_000107_370 |
0.2074368971 |
- |
- |
PREDICTED: uncharacterized protein At2g17340-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_006816_500 |
0.2076254501 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 14 |
Hb_015026_120 |
0.2084766056 |
- |
- |
hypothetical protein TRIUR3_03502 [Triticum urartu] |
| 15 |
Hb_055384_010 |
0.210864878 |
- |
- |
- |
| 16 |
Hb_000500_280 |
0.2124051927 |
- |
- |
Brassinosteroid-regulated protein BRU1 precursor, putative [Ricinus communis] |
| 17 |
Hb_134226_010 |
0.2131222998 |
- |
- |
40S ribosomal protein S21e, putative [Ricinus communis] |
| 18 |
Hb_001150_040 |
0.2133986659 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 19 |
Hb_157854_010 |
0.2135147426 |
- |
- |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 20 |
Hb_003777_270 |
0.2142490414 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |