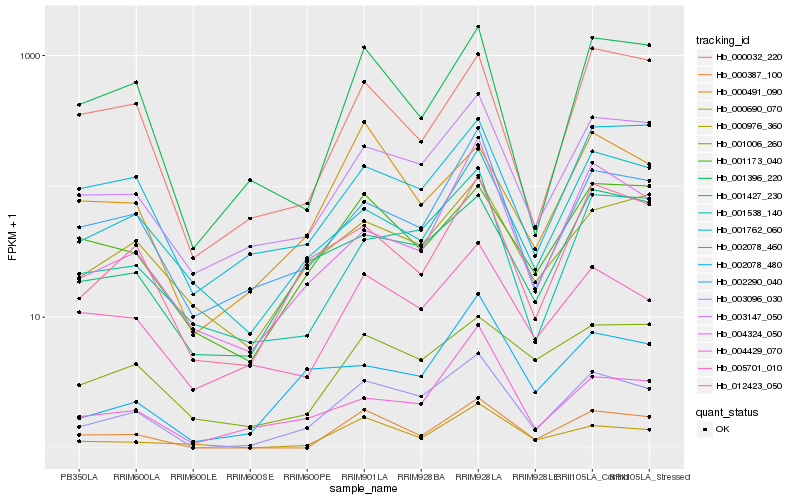
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003096_030 |
0.0 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003147_050 |
0.1342631612 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_012423_050 |
0.1524492455 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 4 |
Hb_005701_010 |
0.1614178855 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001538_140 |
0.1621281015 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 6 |
Hb_002290_040 |
0.1626585557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001427_230 |
0.1646109109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000387_100 |
0.1660728744 |
- |
- |
- |
| 9 |
Hb_004324_050 |
0.1723940663 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 10 |
Hb_000976_360 |
0.1739008409 |
- |
- |
PREDICTED: uncharacterized protein LOC105646237 [Jatropha curcas] |
| 11 |
Hb_002078_480 |
0.1741476816 |
- |
- |
hypothetical protein CICLE_v10033318mg [Citrus clementina] |
| 12 |
Hb_001396_220 |
0.176257122 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 13 |
Hb_001173_040 |
0.1769974541 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 14 |
Hb_000690_070 |
0.1783604259 |
- |
- |
- |
| 15 |
Hb_001006_260 |
0.1804387303 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 16 |
Hb_002078_460 |
0.1815060382 |
- |
- |
- |
| 17 |
Hb_004429_070 |
0.1837763298 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_000491_090 |
0.1838912382 |
- |
- |
hypothetical protein B456_008G219400 [Gossypium raimondii] |
| 19 |
Hb_001762_060 |
0.1854339508 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 20 |
Hb_000032_220 |
0.1900460967 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |