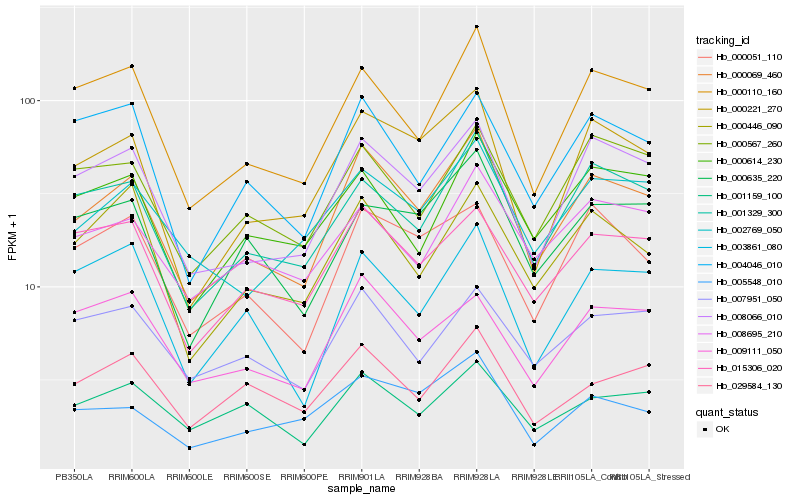
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_460 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 2 |
Hb_008066_010 |
0.0796869974 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |
| 3 |
Hb_000221_270 |
0.0874985789 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 4 |
Hb_000110_160 |
0.0877507593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001329_300 |
0.091879334 |
- |
- |
PREDICTED: L-cysteine desulfhydrase [Jatropha curcas] |
| 6 |
Hb_000051_110 |
0.1036800241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000446_090 |
0.1042474237 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 8 |
Hb_002769_050 |
0.1042501659 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 9 |
Hb_015306_020 |
0.1044857092 |
transcription factor |
TF Family: MYB-related |
PREDICTED: dnaJ homolog subfamily C member 2 [Jatropha curcas] |
| 10 |
Hb_000567_260 |
0.1083274346 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 11 |
Hb_001159_100 |
0.1096516055 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g22760 [Jatropha curcas] |
| 12 |
Hb_004046_010 |
0.1103793203 |
- |
- |
PREDICTED: uncharacterized protein LOC105632478 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005548_010 |
0.1111161763 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 14 |
Hb_029584_130 |
0.111819714 |
- |
- |
PREDICTED: uncharacterized protein LOC105637865 [Jatropha curcas] |
| 15 |
Hb_007951_050 |
0.1118692609 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000635_220 |
0.1121751685 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 17 |
Hb_000614_230 |
0.1123082962 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 18 |
Hb_008695_210 |
0.1127134429 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 19 |
Hb_009111_050 |
0.1135870351 |
transcription factor |
TF Family: Jumonji |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540 [Populus euphratica] |
| 20 |
Hb_003861_080 |
0.1142754734 |
- |
- |
PREDICTED: DNA polymerase kappa isoform X2 [Jatropha curcas] |