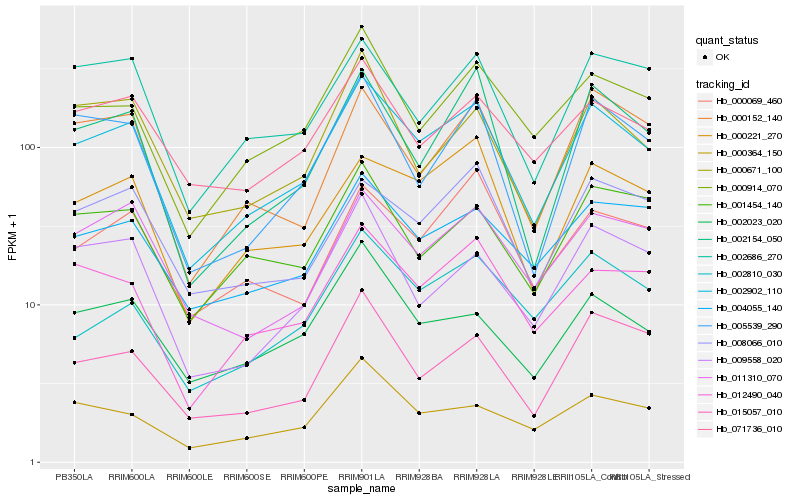
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002902_110 |
0.0 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 2 |
Hb_000221_270 |
0.0998412533 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 3 |
Hb_000914_070 |
0.1013427232 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 8 [Jatropha curcas] |
| 4 |
Hb_012490_040 |
0.1016077587 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 5 |
Hb_002154_050 |
0.1055781738 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_004055_140 |
0.1058715833 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 7 |
Hb_071736_010 |
0.1066725478 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_002686_270 |
0.1080998705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005539_290 |
0.108244275 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_011310_070 |
0.1090226867 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 11 |
Hb_008066_010 |
0.1112775144 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_002023_020 |
0.1126248378 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 13 |
Hb_015057_010 |
0.1126669028 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |
| 14 |
Hb_000152_140 |
0.1127853919 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000671_100 |
0.1161708614 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |
| 16 |
Hb_000069_460 |
0.1165933823 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 17 |
Hb_001454_140 |
0.1175139104 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_009558_020 |
0.1182198159 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002810_030 |
0.1183100152 |
- |
- |
hypothetical protein VITISV_015705 [Vitis vinifera] |
| 20 |
Hb_000364_150 |
0.118341511 |
- |
- |
- |