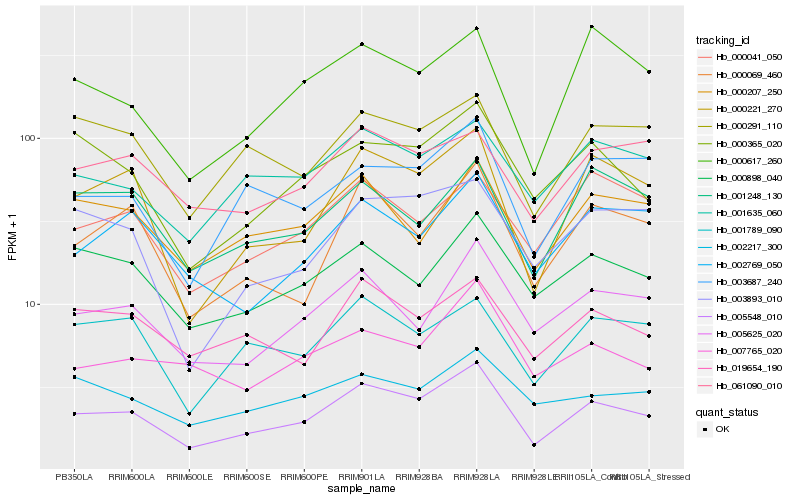
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005548_010 |
0.0 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 2 |
Hb_000221_270 |
0.0934281601 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 3 |
Hb_005625_020 |
0.1103088015 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000069_460 |
0.1111161763 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 5 |
Hb_003687_240 |
0.1130673046 |
- |
- |
PREDICTED: cryptochrome-1 [Jatropha curcas] |
| 6 |
Hb_001635_060 |
0.1142195565 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 7 |
Hb_000365_020 |
0.1148616259 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000898_040 |
0.1171482847 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000207_250 |
0.1185711124 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 10 |
Hb_000617_260 |
0.1198011613 |
rubber biosynthesis |
Gene Name: SRPP2 |
small rubber particle protein [Hevea brasiliensis] |
| 11 |
Hb_019654_190 |
0.1200463574 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001789_090 |
0.1211242296 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |
| 13 |
Hb_000291_110 |
0.1217041326 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 14 |
Hb_003893_010 |
0.1227893533 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 15 |
Hb_002769_050 |
0.1233560293 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 16 |
Hb_007765_020 |
0.1246022189 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 17 |
Hb_002217_300 |
0.1259340991 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 18 |
Hb_001248_130 |
0.1278455148 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 19 |
Hb_061090_010 |
0.1279636984 |
- |
- |
RAN BINDING protein 1 [Populus trichocarpa] |
| 20 |
Hb_000041_050 |
0.1282370732 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |