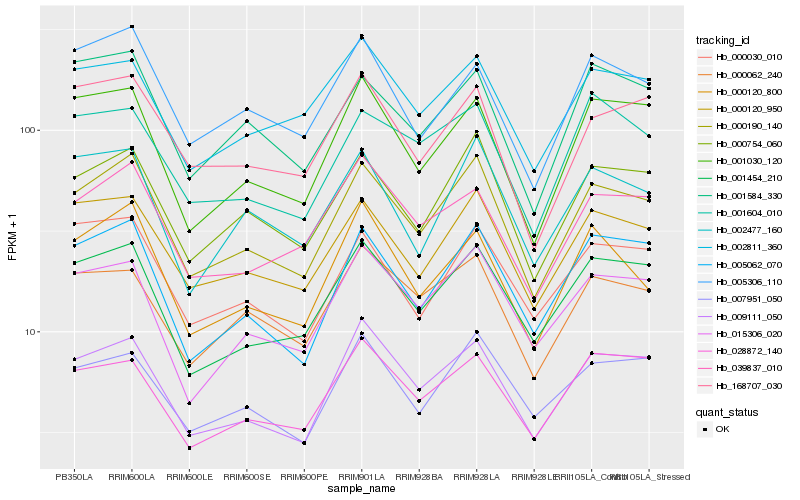
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_140 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000754_060 |
0.0476381997 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000120_950 |
0.0559243863 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 4 |
Hb_009111_050 |
0.060364355 |
transcription factor |
TF Family: Jumonji |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540 [Populus euphratica] |
| 5 |
Hb_001454_210 |
0.0633071478 |
- |
- |
PREDICTED: uncharacterized protein LOC105643435 [Jatropha curcas] |
| 6 |
Hb_005062_070 |
0.063716491 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 7 |
Hb_015306_020 |
0.0717846296 |
transcription factor |
TF Family: MYB-related |
PREDICTED: dnaJ homolog subfamily C member 2 [Jatropha curcas] |
| 8 |
Hb_001030_120 |
0.0735267173 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_007951_050 |
0.0740388065 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000030_010 |
0.0759393188 |
- |
- |
PREDICTED: uncharacterized protein LOC105632627 [Jatropha curcas] |
| 11 |
Hb_000062_240 |
0.0762730438 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 12 |
Hb_002477_160 |
0.0765510596 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 13 |
Hb_039837_010 |
0.0773842791 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 14 |
Hb_000120_800 |
0.0775198092 |
- |
- |
PREDICTED: rab proteins geranylgeranyltransferase component A [Jatropha curcas] |
| 15 |
Hb_028872_140 |
0.0789728238 |
- |
- |
PREDICTED: uncharacterized protein LOC105644767 isoform X1 [Jatropha curcas] |
| 16 |
Hb_168707_030 |
0.079043733 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 17 |
Hb_001584_330 |
0.0791396719 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 18 |
Hb_002811_360 |
0.0805501559 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 19 |
Hb_005306_110 |
0.0807841463 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 20 |
Hb_001604_010 |
0.0821826373 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |