| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
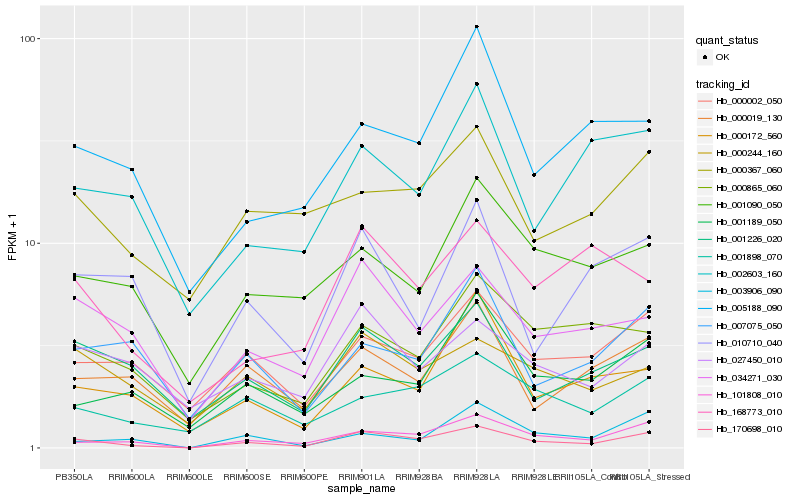
Hb_170698_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 2 |
Hb_101808_010 |
0.1371224216 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 3 |
Hb_034271_030 |
0.1881034387 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001226_020 |
0.191665739 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000002_050 |
0.1920154726 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 6 |
Hb_001898_070 |
0.1940355476 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 7 |
Hb_000244_160 |
0.2029302226 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g24000, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000019_130 |
0.2029987112 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At2g01510 [Jatropha curcas] |
| 9 |
Hb_000172_560 |
0.2041821564 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 10 |
Hb_027450_010 |
0.2069362621 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Populus euphratica] |
| 11 |
Hb_003906_090 |
0.2070041953 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 12 |
Hb_000865_060 |
0.2082083406 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 13 |
Hb_007075_050 |
0.2083056835 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_010710_040 |
0.2110025289 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002603_160 |
0.2123000067 |
- |
- |
Arginyl-tRNA--protein transferase, putative [Ricinus communis] |
| 16 |
Hb_168773_010 |
0.2123563977 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_001090_050 |
0.2129382347 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001189_050 |
0.2129522109 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 19 |
Hb_005188_090 |
0.2134488255 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 20 |
Hb_000367_060 |
0.2135185312 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |