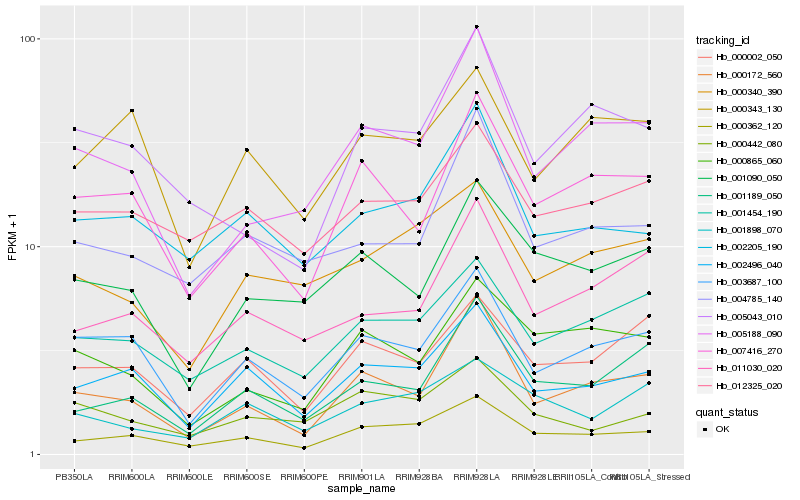
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_120 |
0.0 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 2 |
Hb_002496_040 |
0.0926613799 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002205_190 |
0.1094633344 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 4 |
Hb_001454_190 |
0.1143675569 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 5 |
Hb_012325_020 |
0.1271584707 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000340_390 |
0.1290454603 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 7 |
Hb_007416_270 |
0.1290938432 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 8 |
Hb_003687_100 |
0.1296622292 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 9 |
Hb_000172_560 |
0.1315763304 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 10 |
Hb_000002_050 |
0.132867277 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 11 |
Hb_001898_070 |
0.1345294905 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 12 |
Hb_005188_090 |
0.1346549915 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 13 |
Hb_001189_050 |
0.1348522049 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 14 |
Hb_005043_010 |
0.1367413769 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_000442_080 |
0.1383676056 |
- |
- |
hypothetical protein JCGZ_03373 [Jatropha curcas] |
| 16 |
Hb_004785_140 |
0.1403370196 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_011030_020 |
0.1411390773 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 18 |
Hb_000865_060 |
0.1440242803 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 19 |
Hb_001090_050 |
0.144104075 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000343_130 |
0.1441426043 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |