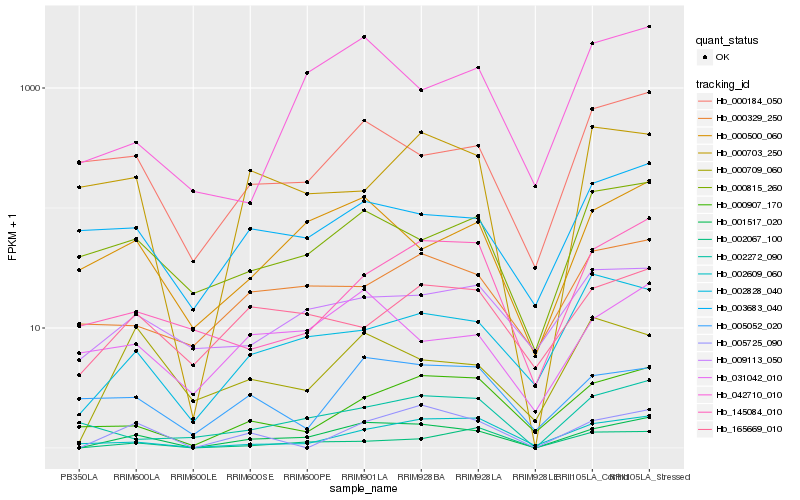
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001517_020 |
0.0 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 2 |
Hb_005725_090 |
0.2122524221 |
- |
- |
PREDICTED: phytosulfokines 3 [Cicer arietinum] |
| 3 |
Hb_000500_060 |
0.2300545017 |
- |
- |
BnaC08g21190D [Brassica napus] |
| 4 |
Hb_002067_100 |
0.237145034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002828_040 |
0.2379197836 |
- |
- |
transmembrane protein, putative [Medicago truncatula] |
| 6 |
Hb_002609_060 |
0.2415765003 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 7 |
Hb_000907_170 |
0.2418263534 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_031042_010 |
0.2479385087 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 9 |
Hb_000184_050 |
0.2493900397 |
- |
- |
NOI, putative [Ricinus communis] |
| 10 |
Hb_042710_010 |
0.2517748101 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 11 |
Hb_165669_010 |
0.2531797613 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000815_260 |
0.2532411199 |
- |
- |
50S ribosomal protein L32pA [Hevea brasiliensis] |
| 13 |
Hb_003683_040 |
0.2539664042 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000709_060 |
0.2540021602 |
- |
- |
PREDICTED: abscisic-aldehyde oxidase-like isoform X5 [Populus euphratica] |
| 15 |
Hb_005052_020 |
0.2556517588 |
- |
- |
- |
| 16 |
Hb_000329_250 |
0.2556529882 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_002272_090 |
0.2571876101 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 18 |
Hb_009113_050 |
0.257800782 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 19 |
Hb_000703_250 |
0.2594796638 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |
| 20 |
Hb_145084_010 |
0.2597283743 |
- |
- |
PREDICTED: uncharacterized protein LOC105646270 [Jatropha curcas] |