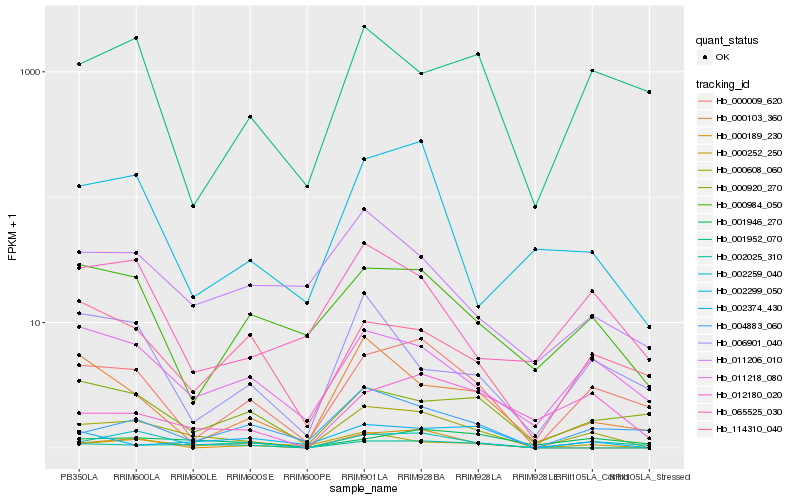
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_000009_620 |
0.2292240697 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000252_250 |
0.2331465214 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_011218_080 |
0.248778383 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 5 |
Hb_002259_040 |
0.2578229744 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002025_310 |
0.2640099711 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002299_050 |
0.2723586956 |
- |
- |
PREDICTED: chalcone--flavonone isomerase isoform X1 [Pyrus x bretschneideri] |
| 8 |
Hb_004883_060 |
0.2744819693 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000103_360 |
0.2757143547 |
- |
- |
- |
| 10 |
Hb_065525_030 |
0.2773561142 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 11 |
Hb_000984_050 |
0.2782837444 |
- |
- |
sucrose transporter 5 [Hevea brasiliensis] |
| 12 |
Hb_012180_020 |
0.2794987046 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |
| 13 |
Hb_011206_010 |
0.2850476558 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 14 |
Hb_006901_040 |
0.2856358384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000189_230 |
0.2880615057 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 16 |
Hb_002374_430 |
0.2896101576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_114310_040 |
0.2899303831 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 18 |
Hb_001946_270 |
0.290968619 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 19 |
Hb_000920_270 |
0.2911227345 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 20 |
Hb_001952_070 |
0.2920647347 |
- |
- |
40S ribosomal protein S6B [Hevea brasiliensis] |