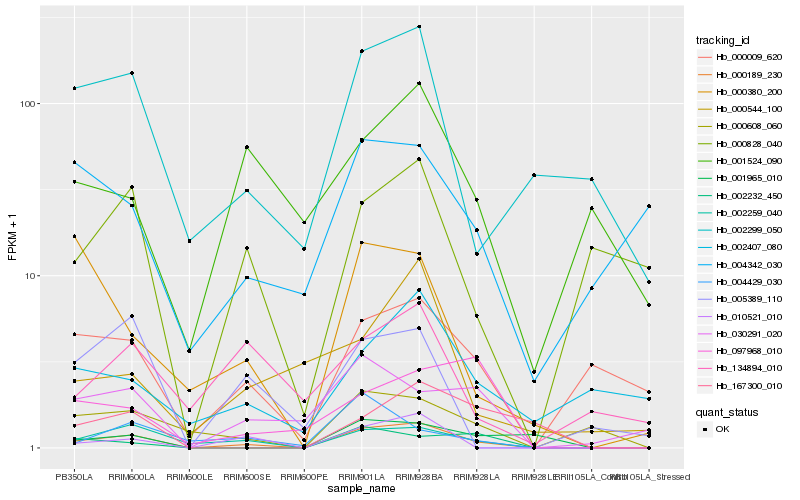
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_230 |
0.0 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 2 |
Hb_002259_040 |
0.2176145732 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_010521_010 |
0.262669578 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 4 |
Hb_000608_060 |
0.2880615057 |
- |
- |
- |
| 5 |
Hb_002299_050 |
0.2904646995 |
- |
- |
PREDICTED: chalcone--flavonone isomerase isoform X1 [Pyrus x bretschneideri] |
| 6 |
Hb_000009_620 |
0.3004320441 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_030291_020 |
0.3118264555 |
- |
- |
PREDICTED: protein argonaute 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002232_450 |
0.312031782 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 9 |
Hb_005389_110 |
0.312480685 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 10 |
Hb_001965_010 |
0.3135816646 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 11 |
Hb_000380_200 |
0.3193484024 |
- |
- |
- |
| 12 |
Hb_000828_040 |
0.3211136267 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 13 |
Hb_167300_010 |
0.3283198124 |
- |
- |
- |
| 14 |
Hb_002407_080 |
0.3344799855 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 15 |
Hb_004429_030 |
0.334824446 |
- |
- |
hypothetical protein POPTR_0010s16560g [Populus trichocarpa] |
| 16 |
Hb_097968_010 |
0.3382278043 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 17 |
Hb_000544_100 |
0.3388933313 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 18 |
Hb_004342_030 |
0.339681636 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 19 |
Hb_001524_090 |
0.3420059512 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 20 |
Hb_134894_010 |
0.3423682726 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |