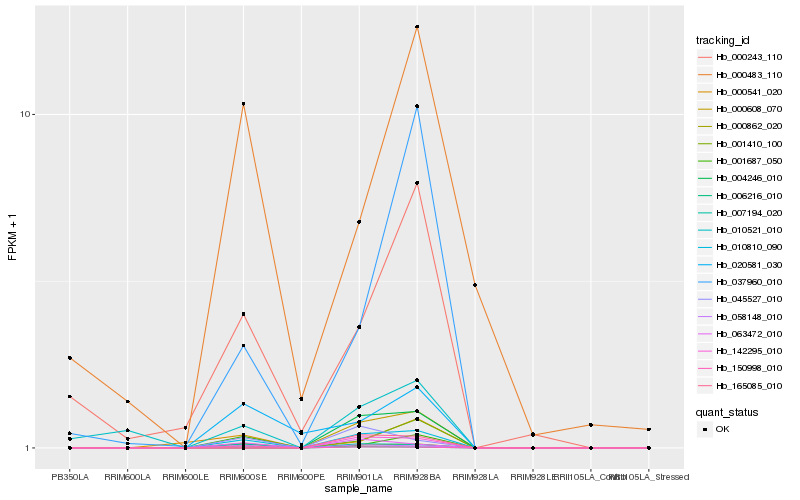
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010810_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_007194_020 |
0.1536443039 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 3 |
Hb_058148_010 |
0.1595833042 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 4 |
Hb_000862_020 |
0.184666404 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 5 |
Hb_063472_010 |
0.21475126 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 6 |
Hb_045527_010 |
0.2402186788 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 7 |
Hb_001410_100 |
0.2481934361 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 8 |
Hb_020581_030 |
0.2766926943 |
- |
- |
- |
| 9 |
Hb_006216_010 |
0.281297071 |
- |
- |
PREDICTED: uncharacterized protein LOC103411650 [Malus domestica] |
| 10 |
Hb_150998_010 |
0.2865962703 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 11 |
Hb_001687_050 |
0.2873207741 |
- |
- |
- |
| 12 |
Hb_010521_010 |
0.2949940475 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 13 |
Hb_000243_110 |
0.2964143828 |
- |
- |
PREDICTED: WAT1-related protein At3g53210-like [Jatropha curcas] |
| 14 |
Hb_000541_020 |
0.3184866139 |
- |
- |
- |
| 15 |
Hb_004246_010 |
0.3302232304 |
- |
- |
- |
| 16 |
Hb_000608_070 |
0.3315755084 |
- |
- |
- |
| 17 |
Hb_037960_010 |
0.332158486 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_165085_010 |
0.3366646537 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 19 |
Hb_142295_010 |
0.337369412 |
- |
- |
- |
| 20 |
Hb_000483_110 |
0.3383973571 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 2, partial [Theobroma cacao] |