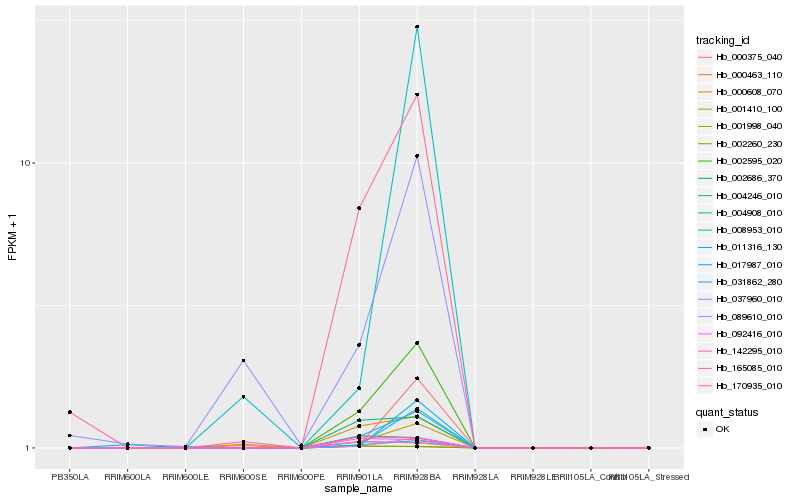
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_280 |
0.0 |
- |
- |
hypothetical protein VITISV_012289 [Vitis vinifera] |
| 2 |
Hb_002595_020 |
0.0145046918 |
- |
- |
PREDICTED: class I heat shock protein, partial [Nicotiana sylvestris] |
| 3 |
Hb_004908_010 |
0.0674175799 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 4 |
Hb_089610_010 |
0.0744036871 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 5 |
Hb_170935_010 |
0.103805752 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Setaria italica] |
| 6 |
Hb_000608_070 |
0.167937172 |
- |
- |
- |
| 7 |
Hb_004246_010 |
0.2234895936 |
- |
- |
- |
| 8 |
Hb_001410_100 |
0.2347761602 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 9 |
Hb_037960_010 |
0.2517371972 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_011316_130 |
0.266629393 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_017987_010 |
0.2741154673 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 3-like isoform X1 [Populus euphratica] |
| 12 |
Hb_165085_010 |
0.2751759425 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 13 |
Hb_142295_010 |
0.2786959282 |
- |
- |
- |
| 14 |
Hb_000375_040 |
0.2836299999 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 15 |
Hb_092416_010 |
0.284574071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638802 [Jatropha curcas] |
| 16 |
Hb_002686_370 |
0.2850507667 |
- |
- |
PREDICTED: uncharacterized protein LOC102628436 [Citrus sinensis] |
| 17 |
Hb_000463_110 |
0.2852742964 |
- |
- |
- |
| 18 |
Hb_008953_010 |
0.2902059311 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X2 [Solanum lycopersicum] |
| 19 |
Hb_002260_230 |
0.2930894001 |
- |
- |
hypothetical protein SORBIDRAFT_05g006180 [Sorghum bicolor] |
| 20 |
Hb_001998_040 |
0.2935880996 |
- |
- |
ATP binding protein, putative [Ricinus communis] |