| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
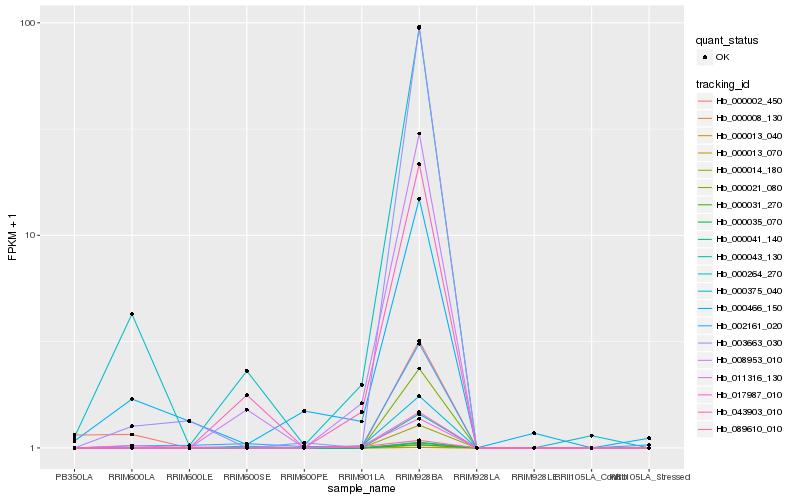
Hb_011316_130 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000264_270 |
0.1585668355 |
transcription factor |
TF Family: HSF |
- |
| 3 |
Hb_000466_150 |
0.2135432338 |
- |
- |
hypothetical protein JCGZ_02123 [Jatropha curcas] |
| 4 |
Hb_017987_010 |
0.2154374564 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 3-like isoform X1 [Populus euphratica] |
| 5 |
Hb_008953_010 |
0.2180886781 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X2 [Solanum lycopersicum] |
| 6 |
Hb_089610_010 |
0.2197000013 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 7 |
Hb_000375_040 |
0.2208390195 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 8 |
Hb_043903_010 |
0.2359107371 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Tarenaya hassleriana] |
| 9 |
Hb_003663_030 |
0.239019113 |
- |
- |
PREDICTED: early nodulin-93-like [Jatropha curcas] |
| 10 |
Hb_002161_020 |
0.2403725315 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 11 |
Hb_000002_450 |
0.2404175134 |
- |
- |
hypothetical protein JCGZ_02285 [Jatropha curcas] |
| 12 |
Hb_000008_130 |
0.2494644058 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000013_040 |
0.2494644058 |
- |
- |
- |
| 14 |
Hb_000013_070 |
0.2494644058 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_000014_180 |
0.2494644058 |
- |
- |
PREDICTED: endo-1,4-beta-xylanase D-like [Fragaria vesca subsp. vesca] |
| 16 |
Hb_000021_080 |
0.2494644058 |
- |
- |
- |
| 17 |
Hb_000031_270 |
0.2494644058 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000035_070 |
0.2494644058 |
- |
- |
PREDICTED: uncharacterized protein LOC104814513 [Tarenaya hassleriana] |
| 19 |
Hb_000041_140 |
0.2494644058 |
- |
- |
- |
| 20 |
Hb_000043_130 |
0.2494644058 |
- |
- |
- |