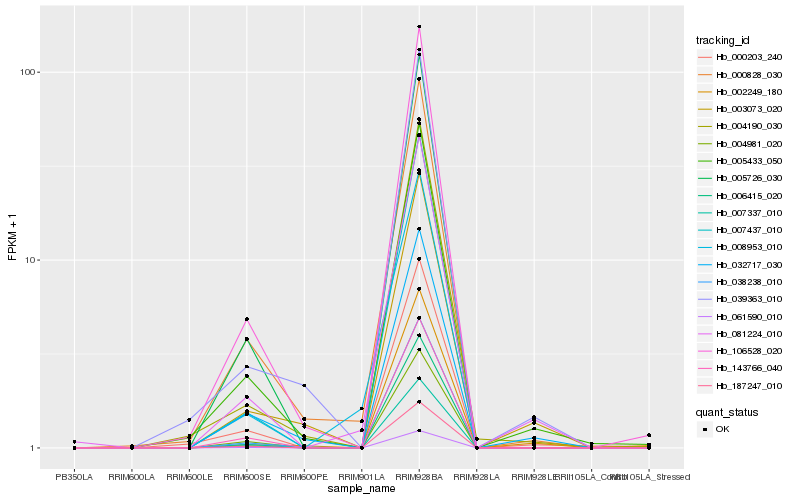
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081224_010 |
0.0 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 2 |
Hb_000828_030 |
0.0848646928 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 3 |
Hb_005433_050 |
0.0871143461 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 4 |
Hb_005726_030 |
0.0897462431 |
- |
- |
hypothetical protein CISIN_1g020511mg [Citrus sinensis] |
| 5 |
Hb_061590_010 |
0.0898940125 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 6 |
Hb_187247_010 |
0.0923349077 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 7 |
Hb_007437_010 |
0.0926758335 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_008953_010 |
0.0941343478 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X2 [Solanum lycopersicum] |
| 9 |
Hb_007337_010 |
0.0958997865 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 10 |
Hb_000203_240 |
0.0960034767 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_006415_020 |
0.0960063749 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Jatropha curcas] |
| 12 |
Hb_143766_040 |
0.0964348116 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 13 |
Hb_032717_030 |
0.0966942679 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 14 |
Hb_003073_020 |
0.0967613232 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_038238_010 |
0.0968617207 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 16 |
Hb_004981_020 |
0.0975655528 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |
| 17 |
Hb_106528_020 |
0.0984896474 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 18 |
Hb_002249_180 |
0.1008652233 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_004190_030 |
0.1015926462 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_039363_010 |
0.1029248414 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |