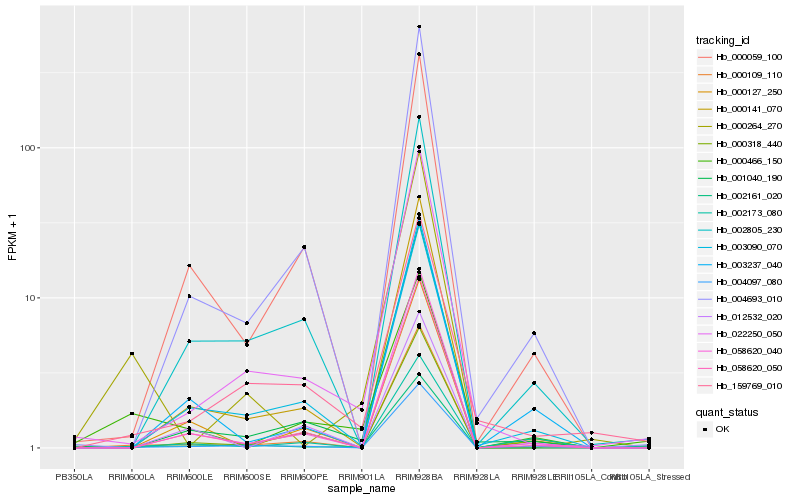
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000466_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_02123 [Jatropha curcas] |
| 2 |
Hb_002161_020 |
0.1816634751 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 3 |
Hb_002173_080 |
0.1912454104 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000109_110 |
0.1941398356 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_004097_080 |
0.196511521 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 6 |
Hb_001040_190 |
0.1986067847 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 7 |
Hb_000059_100 |
0.1998426677 |
- |
- |
Basic blue protein, putative [Ricinus communis] |
| 8 |
Hb_000264_270 |
0.2008113016 |
transcription factor |
TF Family: HSF |
- |
| 9 |
Hb_004693_010 |
0.2028520303 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 10 |
Hb_000318_440 |
0.2037777233 |
- |
- |
PREDICTED: allene oxide synthase 2-like [Jatropha curcas] |
| 11 |
Hb_000127_250 |
0.2044335071 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 12 |
Hb_022250_050 |
0.2047323653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_058620_050 |
0.2057694092 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 14 |
Hb_003090_070 |
0.2063414771 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 15 |
Hb_058620_040 |
0.2067924981 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_000141_070 |
0.2075823886 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_003237_040 |
0.2102021967 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_002805_230 |
0.2104514847 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 19 |
Hb_159769_010 |
0.2105349594 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 20 |
Hb_012532_020 |
0.2108515044 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |