| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
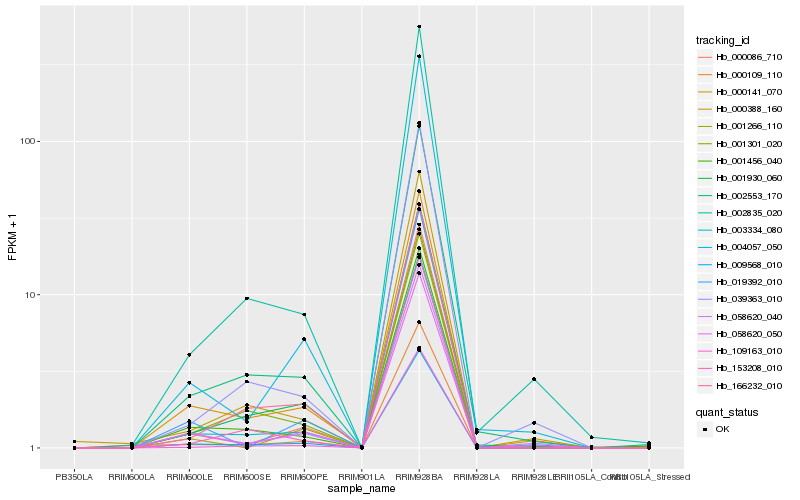
Hb_000109_110 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000086_710 |
0.0722874562 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 3 |
Hb_009568_010 |
0.07462736 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 4 |
Hb_003334_080 |
0.0763738818 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 5 |
Hb_109163_010 |
0.0767500022 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 6 |
Hb_001930_060 |
0.0796708613 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 7 |
Hb_002553_170 |
0.0804880633 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 8 |
Hb_000388_160 |
0.0807224756 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 9 |
Hb_058620_050 |
0.0823887805 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 10 |
Hb_000141_070 |
0.0829297601 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_166232_010 |
0.083781805 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |
| 12 |
Hb_002835_020 |
0.0848625293 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 13 |
Hb_058620_040 |
0.084878951 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 14 |
Hb_004057_050 |
0.0853382395 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 15 |
Hb_019392_010 |
0.0878849129 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 16 |
Hb_001456_040 |
0.0882574949 |
- |
- |
NAD dependent epimerase/dehydratase, putative [Ricinus communis] |
| 17 |
Hb_001266_110 |
0.0886393964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_039363_010 |
0.0904037927 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_153208_010 |
0.0907233948 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_001301_020 |
0.0912588728 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |