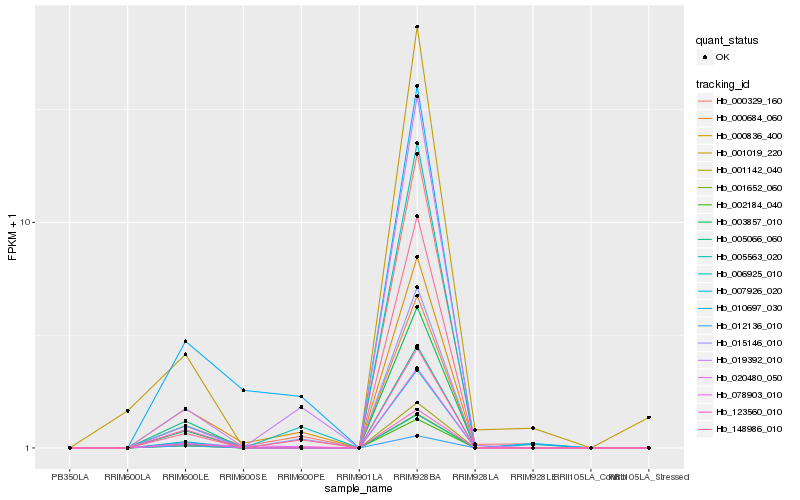
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003857_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001142_040 |
0.0065740492 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 3 |
Hb_002184_040 |
0.0080758968 |
transcription factor |
TF Family: bHLH |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_007926_020 |
0.0085671946 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 5 |
Hb_001652_060 |
0.0375595375 |
- |
- |
PREDICTED: uncharacterized protein LOC103404034 [Malus domestica] |
| 6 |
Hb_078903_010 |
0.057467404 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 7 |
Hb_123560_010 |
0.0753621139 |
- |
- |
hypothetical protein JCGZ_12463 [Jatropha curcas] |
| 8 |
Hb_005563_020 |
0.0972734159 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 9 |
Hb_015146_010 |
0.1037610627 |
- |
- |
- |
| 10 |
Hb_148986_010 |
0.1137316127 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000684_060 |
0.1145523075 |
- |
- |
hypothetical protein POPTR_0006s13170g [Populus trichocarpa] |
| 12 |
Hb_001019_220 |
0.1192008322 |
- |
- |
hypothetical protein POPTR_0004s19470g [Populus trichocarpa] |
| 13 |
Hb_000329_160 |
0.1277638185 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_020480_050 |
0.1278650446 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 15 |
Hb_012136_010 |
0.1292413322 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_005066_060 |
0.1297490755 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 17 |
Hb_019392_010 |
0.1330706873 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 18 |
Hb_006925_010 |
0.1338293024 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 19 |
Hb_000836_400 |
0.1344057189 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 20 |
Hb_010697_030 |
0.135708998 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |