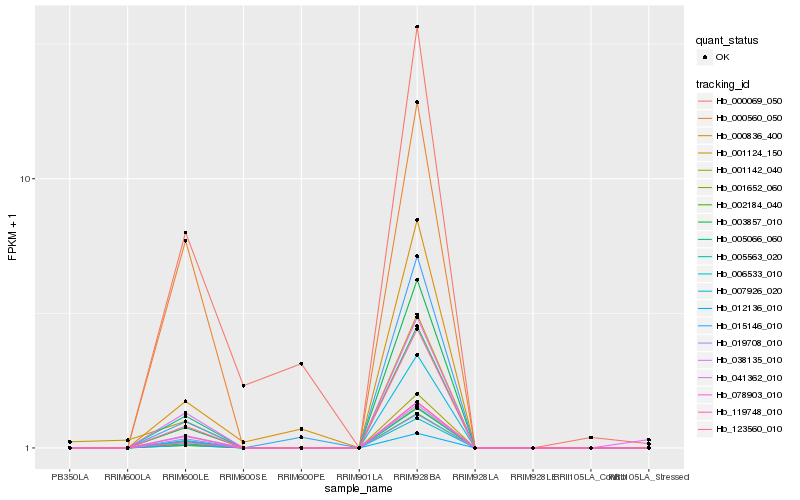
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123560_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_12463 [Jatropha curcas] |
| 2 |
Hb_078903_010 |
0.018046337 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 3 |
Hb_005563_020 |
0.0221935376 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 4 |
Hb_001652_060 |
0.0380237042 |
- |
- |
PREDICTED: uncharacterized protein LOC103404034 [Malus domestica] |
| 5 |
Hb_012136_010 |
0.054726881 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 6 |
Hb_005066_060 |
0.0552448409 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 7 |
Hb_002184_040 |
0.0673780835 |
transcription factor |
TF Family: bHLH |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001142_040 |
0.0688648386 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 9 |
Hb_006533_010 |
0.0733239496 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 10 |
Hb_003857_010 |
0.0753621139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007926_020 |
0.0838019279 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 12 |
Hb_119748_010 |
0.0920057907 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_019708_010 |
0.1099976298 |
- |
- |
- |
| 14 |
Hb_038135_010 |
0.1140010255 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 15 |
Hb_000560_050 |
0.1264033225 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 16 |
Hb_015146_010 |
0.1268973596 |
- |
- |
- |
| 17 |
Hb_041362_010 |
0.1339905483 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 18 |
Hb_000836_400 |
0.1370049562 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 19 |
Hb_000069_050 |
0.1533062736 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 20 |
Hb_001124_150 |
0.1581126344 |
- |
- |
- |