| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
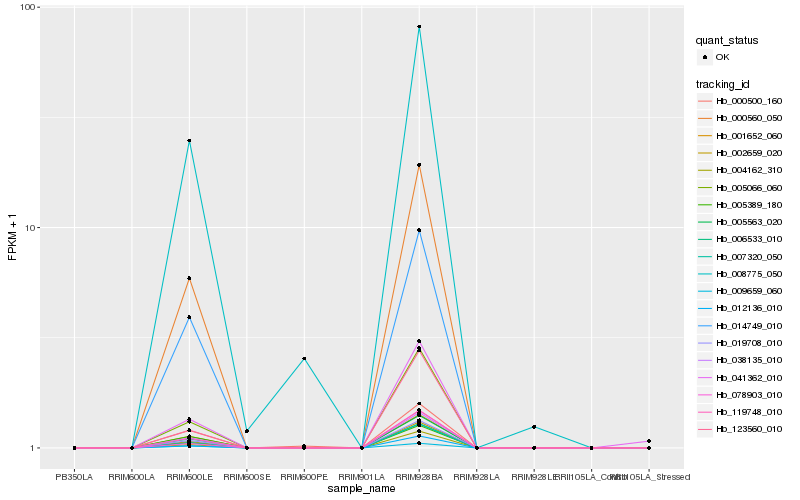
Hb_119748_010 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_019708_010 |
0.0181683388 |
- |
- |
- |
| 3 |
Hb_006533_010 |
0.0188133736 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 4 |
Hb_038135_010 |
0.0222170901 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 5 |
Hb_000560_050 |
0.0347729256 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 6 |
Hb_005066_060 |
0.0369633198 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 7 |
Hb_012136_010 |
0.0374824243 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_005563_020 |
0.0699776392 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 9 |
Hb_014749_010 |
0.0746489329 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 10 |
Hb_007320_050 |
0.0822029337 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 11 |
Hb_004162_310 |
0.088519929 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_123560_010 |
0.0920057907 |
- |
- |
hypothetical protein JCGZ_12463 [Jatropha curcas] |
| 13 |
Hb_078903_010 |
0.1098212519 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 14 |
Hb_008775_050 |
0.1099167175 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 15 |
Hb_002659_020 |
0.1134365898 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 16 |
Hb_009659_060 |
0.1154536824 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 17 |
Hb_041362_010 |
0.1284156085 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 18 |
Hb_001652_060 |
0.1294295293 |
- |
- |
PREDICTED: uncharacterized protein LOC103404034 [Malus domestica] |
| 19 |
Hb_000500_160 |
0.1339241781 |
- |
- |
hypothetical protein CISIN_1g039640mg, partial [Citrus sinensis] |
| 20 |
Hb_005389_180 |
0.1429942749 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |