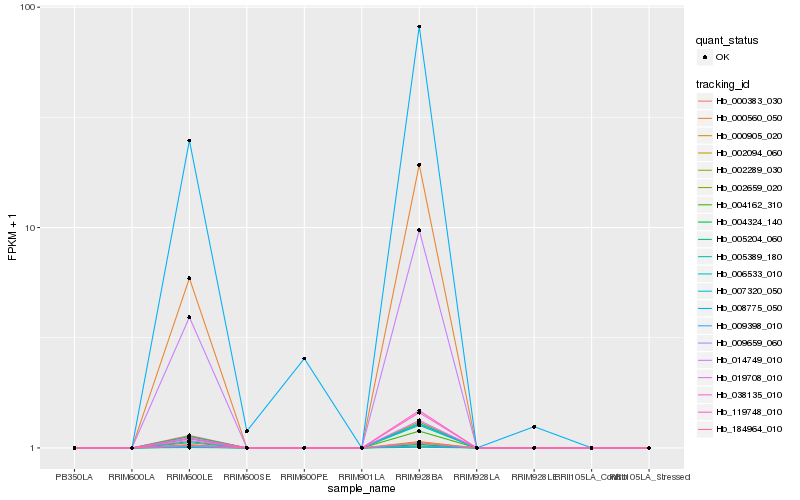
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014749_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 2 |
Hb_007320_050 |
0.0075836215 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 3 |
Hb_004162_310 |
0.0139290009 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 4 |
Hb_002659_020 |
0.038987826 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 5 |
Hb_000560_050 |
0.0399472858 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 6 |
Hb_009659_060 |
0.0410184289 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 7 |
Hb_038135_010 |
0.0524929535 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 8 |
Hb_019708_010 |
0.0565347838 |
- |
- |
- |
| 9 |
Hb_005389_180 |
0.0687710667 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 10 |
Hb_000383_030 |
0.0707081566 |
- |
- |
- |
| 11 |
Hb_119748_010 |
0.0746489329 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_000905_020 |
0.0802410824 |
- |
- |
PREDICTED: cytochrome P450 93A2-like [Jatropha curcas] |
| 13 |
Hb_004324_140 |
0.083400465 |
- |
- |
PREDICTED: uncharacterized protein LOC105648358 [Jatropha curcas] |
| 14 |
Hb_184964_010 |
0.0841208592 |
- |
- |
- |
| 15 |
Hb_002094_060 |
0.0850840301 |
- |
- |
- |
| 16 |
Hb_005204_060 |
0.0862958919 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_006533_010 |
0.0933635819 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 18 |
Hb_002289_030 |
0.0946009456 |
- |
- |
PREDICTED: uncharacterized protein LOC103957017 [Pyrus x bretschneideri] |
| 19 |
Hb_009398_010 |
0.095900691 |
- |
- |
Putative pectate lyase 2 [Glycine soja] |
| 20 |
Hb_008775_050 |
0.0994871827 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |