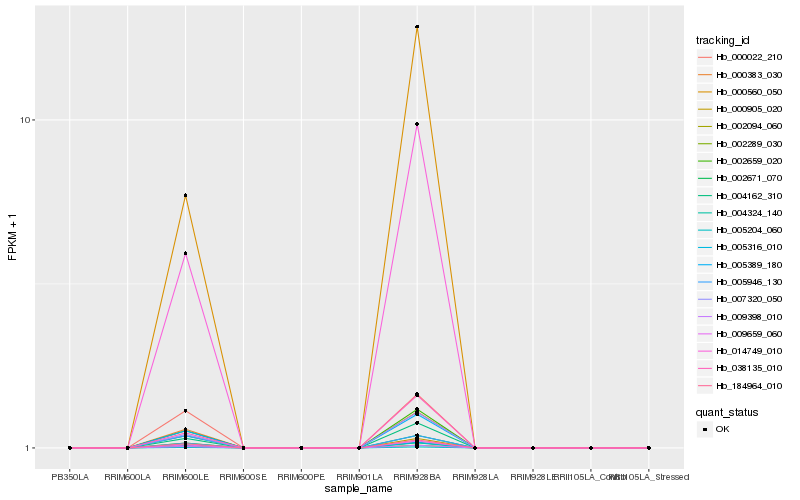
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_020 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 93A2-like [Jatropha curcas] |
| 2 |
Hb_004324_140 |
0.0031692592 |
- |
- |
PREDICTED: uncharacterized protein LOC105648358 [Jatropha curcas] |
| 3 |
Hb_184964_010 |
0.0038920005 |
- |
- |
- |
| 4 |
Hb_002094_060 |
0.0048583642 |
- |
- |
- |
| 5 |
Hb_005204_060 |
0.0060743321 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000383_030 |
0.0095585528 |
- |
- |
- |
| 7 |
Hb_005389_180 |
0.0115000712 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 8 |
Hb_002289_030 |
0.0144101607 |
- |
- |
PREDICTED: uncharacterized protein LOC103957017 [Pyrus x bretschneideri] |
| 9 |
Hb_009398_010 |
0.0157151338 |
- |
- |
Putative pectate lyase 2 [Glycine soja] |
| 10 |
Hb_009659_060 |
0.039285958 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 11 |
Hb_002659_020 |
0.0413166945 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 12 |
Hb_000022_210 |
0.0509326602 |
- |
- |
PREDICTED: uncharacterized protein LOC105638395 [Jatropha curcas] |
| 13 |
Hb_004162_310 |
0.0663496322 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 14 |
Hb_007320_050 |
0.0726800377 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 15 |
Hb_014749_010 |
0.0802410824 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 16 |
Hb_005316_010 |
0.1081390931 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000560_050 |
0.1199793361 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 18 |
Hb_038135_010 |
0.1324256068 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 19 |
Hb_002671_070 |
0.133279877 |
- |
- |
hypothetical protein POPTR_0006s05220g, partial [Populus trichocarpa] |
| 20 |
Hb_005946_130 |
0.133279877 |
- |
- |
- |