| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
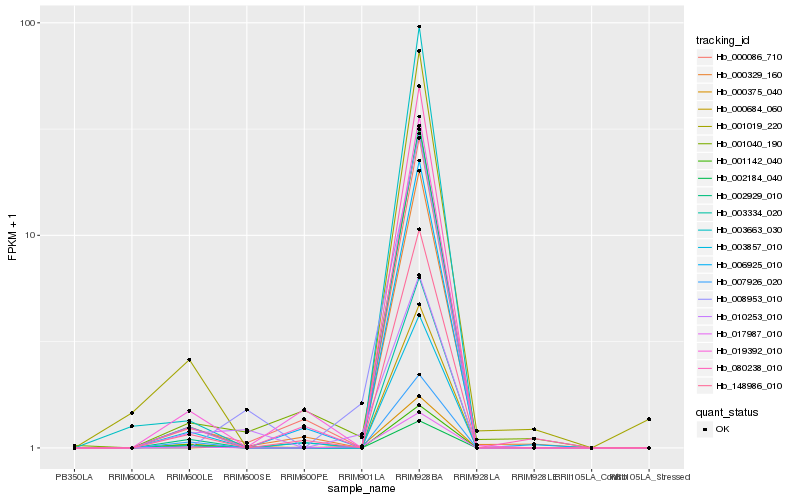
Hb_017987_010 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 3-like isoform X1 [Populus euphratica] |
| 2 |
Hb_000684_060 |
0.1322479261 |
- |
- |
hypothetical protein POPTR_0006s13170g [Populus trichocarpa] |
| 3 |
Hb_010253_010 |
0.1373811146 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Amborella trichopoda] |
| 4 |
Hb_007926_020 |
0.1388823548 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 5 |
Hb_148986_010 |
0.140225303 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_008953_010 |
0.1418105302 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 isoform X2 [Solanum lycopersicum] |
| 7 |
Hb_003857_010 |
0.1429745154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002929_010 |
0.1452915805 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 9 |
Hb_001142_040 |
0.146347418 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 10 |
Hb_000329_160 |
0.1469751408 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002184_040 |
0.1471445404 |
transcription factor |
TF Family: bHLH |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_003663_030 |
0.1490494266 |
- |
- |
PREDICTED: early nodulin-93-like [Jatropha curcas] |
| 13 |
Hb_006925_010 |
0.149949307 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 14 |
Hb_080238_010 |
0.1518789033 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_001019_220 |
0.1523097205 |
- |
- |
hypothetical protein POPTR_0004s19470g [Populus trichocarpa] |
| 16 |
Hb_000375_040 |
0.1524382214 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 17 |
Hb_019392_010 |
0.1526619856 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 18 |
Hb_001040_190 |
0.1558170755 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 19 |
Hb_003334_020 |
0.1566628255 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 20 |
Hb_000086_710 |
0.1573597419 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |