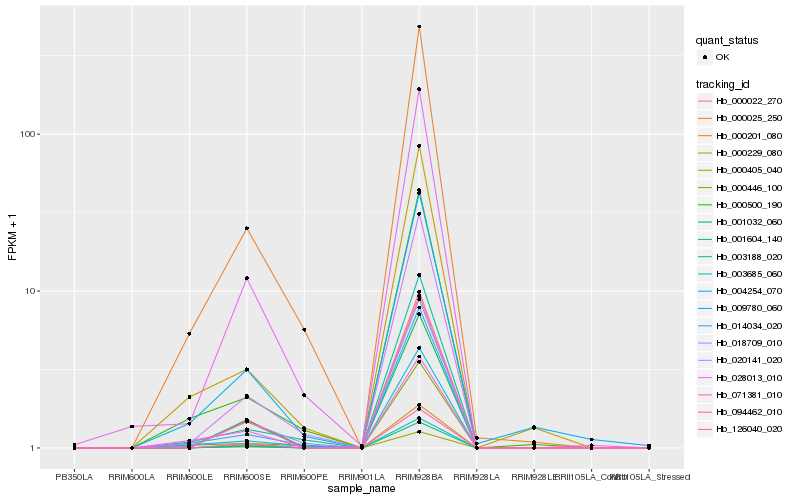
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000405_040 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_014034_020 |
0.0770016597 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 3 |
Hb_000025_250 |
0.0770885533 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 4 |
Hb_071381_010 |
0.0779134577 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 5 |
Hb_001032_060 |
0.078970925 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 6 |
Hb_004254_070 |
0.0821093467 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 7 |
Hb_000229_080 |
0.089638357 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 8 |
Hb_000500_190 |
0.0906527476 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 9 |
Hb_000201_080 |
0.0919140814 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001604_140 |
0.091942818 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 11 |
Hb_003685_060 |
0.0920753776 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |
| 12 |
Hb_126040_020 |
0.0921452703 |
- |
- |
- |
| 13 |
Hb_000022_270 |
0.0922685317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_018709_010 |
0.0927054677 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 15 |
Hb_028013_010 |
0.0935794584 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_009780_060 |
0.0971258349 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_020141_020 |
0.097679399 |
- |
- |
- |
| 18 |
Hb_000446_100 |
0.0980017653 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 19 |
Hb_094462_010 |
0.100818261 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 20 |
Hb_003188_020 |
0.1009385665 |
- |
- |
chitinase, putative [Ricinus communis] |