| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
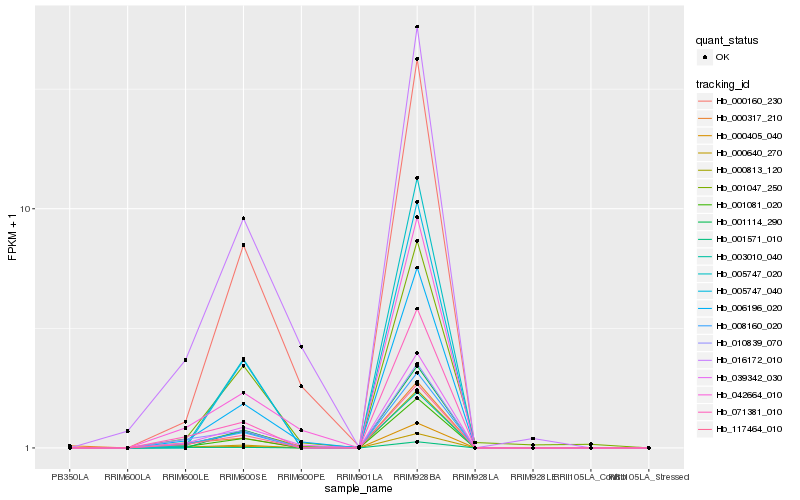
Hb_008160_020 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_117464_010 |
0.0066254961 |
- |
- |
PREDICTED: flavone 3'-O-methyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_001081_020 |
0.0378745486 |
- |
- |
Cytochrome P450 76C2 [Triticum urartu] |
| 4 |
Hb_000640_270 |
0.0385176728 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |
| 5 |
Hb_071381_010 |
0.0536954604 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 6 |
Hb_010839_070 |
0.0541846275 |
- |
- |
uncharacterized protein LOC100810725 [Glycine max] |
| 7 |
Hb_039342_030 |
0.0891644345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006196_020 |
0.0992962954 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 9 |
Hb_001571_010 |
0.1022865982 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 10 |
Hb_000317_210 |
0.1135629362 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39 [Jatropha curcas] |
| 11 |
Hb_000813_120 |
0.1147808816 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 12 |
Hb_005747_020 |
0.1198097792 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |
| 13 |
Hb_000405_040 |
0.1202205808 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_003010_040 |
0.1224600007 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 15 |
Hb_016172_010 |
0.1234684284 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 16 |
Hb_000160_230 |
0.1246793968 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 17 |
Hb_001047_250 |
0.1249815042 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_042664_010 |
0.1250368096 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 19 |
Hb_001114_290 |
0.1266975242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005747_040 |
0.1272850529 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |