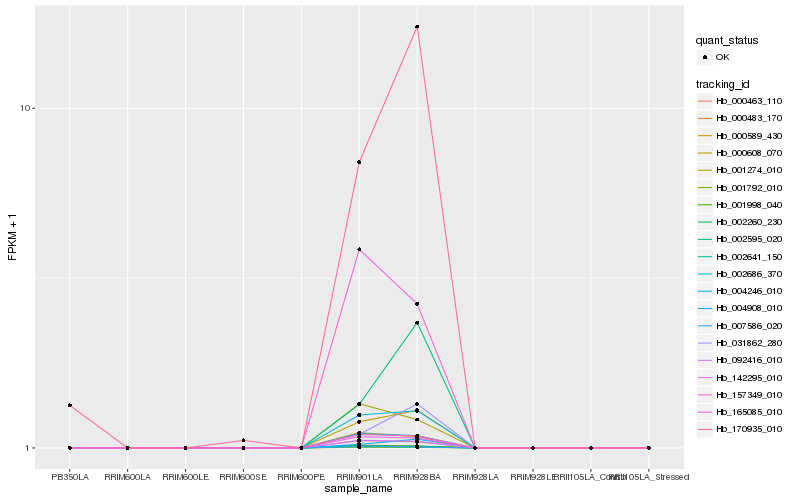
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_004246_010 |
0.0564156703 |
- |
- |
- |
| 3 |
Hb_004908_010 |
0.1010681964 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 4 |
Hb_165085_010 |
0.1092113028 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 5 |
Hb_142295_010 |
0.1128170149 |
- |
- |
- |
| 6 |
Hb_092416_010 |
0.1188411569 |
- |
- |
PREDICTED: uncharacterized protein LOC105638802 [Jatropha curcas] |
| 7 |
Hb_002686_370 |
0.1193298481 |
- |
- |
PREDICTED: uncharacterized protein LOC102628436 [Citrus sinensis] |
| 8 |
Hb_000463_110 |
0.1195590107 |
- |
- |
- |
| 9 |
Hb_002260_230 |
0.1275742704 |
- |
- |
hypothetical protein SORBIDRAFT_05g006180 [Sorghum bicolor] |
| 10 |
Hb_001998_040 |
0.1280859543 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_002641_150 |
0.1291547871 |
- |
- |
PREDICTED: stachyose synthase-like [Citrus sinensis] |
| 12 |
Hb_001792_010 |
0.1299134303 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 13 |
Hb_170935_010 |
0.1521078933 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Setaria italica] |
| 14 |
Hb_000589_430 |
0.1529178094 |
- |
- |
hypothetical protein JCGZ_01402 [Jatropha curcas] |
| 15 |
Hb_000483_170 |
0.1652227614 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 16 |
Hb_031862_280 |
0.167937172 |
- |
- |
hypothetical protein VITISV_012289 [Vitis vinifera] |
| 17 |
Hb_002595_020 |
0.1822058896 |
- |
- |
PREDICTED: class I heat shock protein, partial [Nicotiana sylvestris] |
| 18 |
Hb_001274_010 |
0.1892629695 |
- |
- |
- |
| 19 |
Hb_157349_010 |
0.1980603802 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 20 |
Hb_007586_020 |
0.2218945908 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |