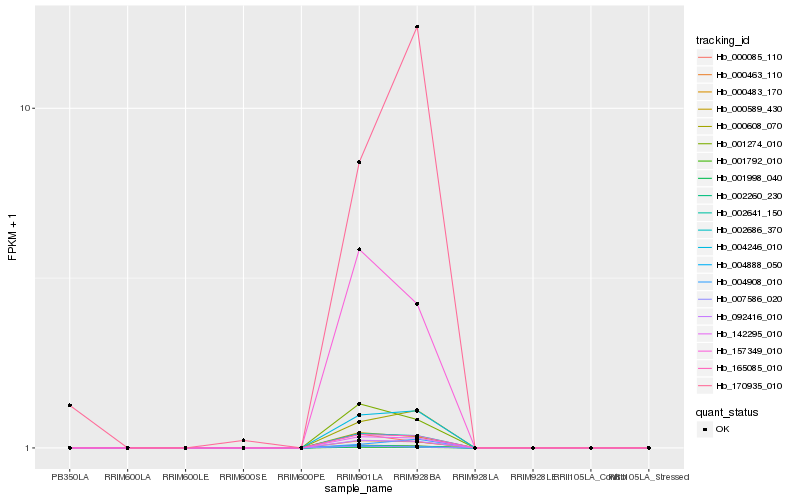
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004246_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_165085_010 |
0.0529097739 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 3 |
Hb_000608_070 |
0.0564156703 |
- |
- |
- |
| 4 |
Hb_142295_010 |
0.0565272182 |
- |
- |
- |
| 5 |
Hb_092416_010 |
0.0625720813 |
- |
- |
PREDICTED: uncharacterized protein LOC105638802 [Jatropha curcas] |
| 6 |
Hb_002686_370 |
0.063062515 |
- |
- |
PREDICTED: uncharacterized protein LOC102628436 [Citrus sinensis] |
| 7 |
Hb_000463_110 |
0.0632924978 |
- |
- |
- |
| 8 |
Hb_002260_230 |
0.0713377235 |
- |
- |
hypothetical protein SORBIDRAFT_05g006180 [Sorghum bicolor] |
| 9 |
Hb_001998_040 |
0.0718514049 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_002641_150 |
0.0729244426 |
- |
- |
PREDICTED: stachyose synthase-like [Citrus sinensis] |
| 11 |
Hb_001792_010 |
0.0736860972 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 12 |
Hb_000589_430 |
0.096792412 |
- |
- |
hypothetical protein JCGZ_01402 [Jatropha curcas] |
| 13 |
Hb_000483_170 |
0.1091603853 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 14 |
Hb_001274_010 |
0.1333410673 |
- |
- |
- |
| 15 |
Hb_157349_010 |
0.1421957086 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 16 |
Hb_004908_010 |
0.1571373345 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 17 |
Hb_007586_020 |
0.1662010292 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 18 |
Hb_004888_050 |
0.1677954083 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_170935_010 |
0.1991993075 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Setaria italica] |
| 20 |
Hb_000085_110 |
0.2042204382 |
- |
- |
hypothetical protein JCGZ_00957 [Jatropha curcas] |