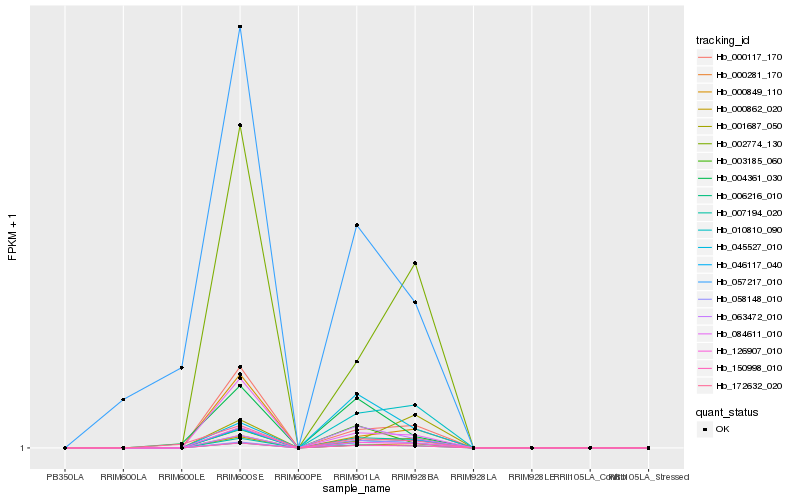
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006216_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103411650 [Malus domestica] |
| 2 |
Hb_150998_010 |
0.0157592648 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 3 |
Hb_000862_020 |
0.1258226387 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 4 |
Hb_046117_040 |
0.1286069141 |
- |
- |
hypothetical protein JCGZ_07941 [Jatropha curcas] |
| 5 |
Hb_007194_020 |
0.1364167378 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 6 |
Hb_126907_010 |
0.1534425367 |
- |
- |
- |
| 7 |
Hb_058148_010 |
0.1592638916 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 8 |
Hb_000281_170 |
0.162573676 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_000117_170 |
0.1741100743 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 10 |
Hb_002774_130 |
0.1878289444 |
- |
- |
- |
| 11 |
Hb_003185_060 |
0.2012298094 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Amborella trichopoda] |
| 12 |
Hb_084611_010 |
0.2030337524 |
- |
- |
PREDICTED: uncharacterized protein LOC104249812 [Nicotiana sylvestris] |
| 13 |
Hb_063472_010 |
0.2052290103 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 14 |
Hb_004361_030 |
0.2059103463 |
- |
- |
PREDICTED: uncharacterized protein LOC104239397 isoform X1 [Nicotiana sylvestris] |
| 15 |
Hb_172632_020 |
0.2217931843 |
- |
- |
- |
| 16 |
Hb_000849_110 |
0.22267456 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 17 |
Hb_045527_010 |
0.2498946185 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 18 |
Hb_057217_010 |
0.2511050077 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 19 |
Hb_001687_050 |
0.2773689565 |
- |
- |
- |
| 20 |
Hb_010810_090 |
0.281297071 |
- |
- |
- |