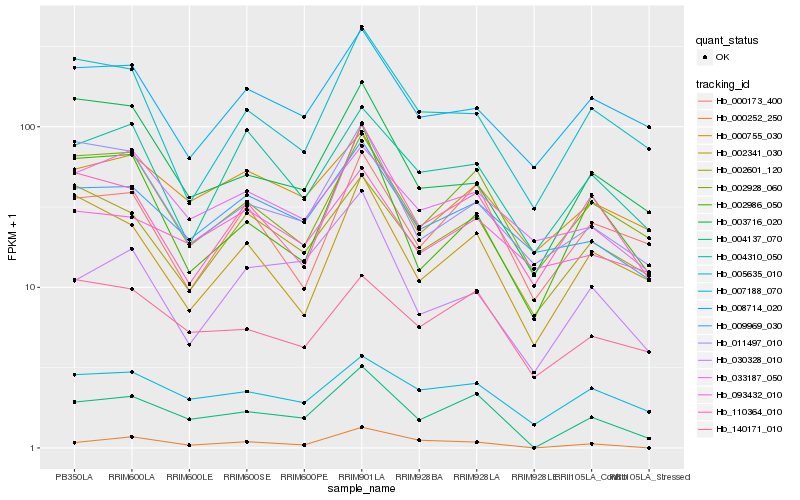
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004137_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_002928_060 |
0.1663068338 |
- |
- |
PREDICTED: pre-rRNA-processing protein TSR1 homolog [Jatropha curcas] |
| 3 |
Hb_110364_010 |
0.1694095936 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 4 |
Hb_007188_070 |
0.1716814351 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 5 |
Hb_000252_250 |
0.1723988395 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_009969_030 |
0.1724420941 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002601_120 |
0.1756069573 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 8 |
Hb_140171_010 |
0.1770049866 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 9 |
Hb_030328_010 |
0.1770778374 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 10 |
Hb_000755_030 |
0.179874046 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 11 |
Hb_000173_400 |
0.1801608695 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |
| 12 |
Hb_002341_030 |
0.1808684402 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 13 |
Hb_011497_010 |
0.1853582039 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 14 |
Hb_002986_050 |
0.1862563967 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 15 |
Hb_004310_050 |
0.1873538446 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 16 |
Hb_005635_010 |
0.188607946 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_003716_020 |
0.1886538964 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 18 |
Hb_033187_050 |
0.1894491316 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 19 |
Hb_093432_010 |
0.190381196 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 20 |
Hb_008714_020 |
0.1904337958 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |