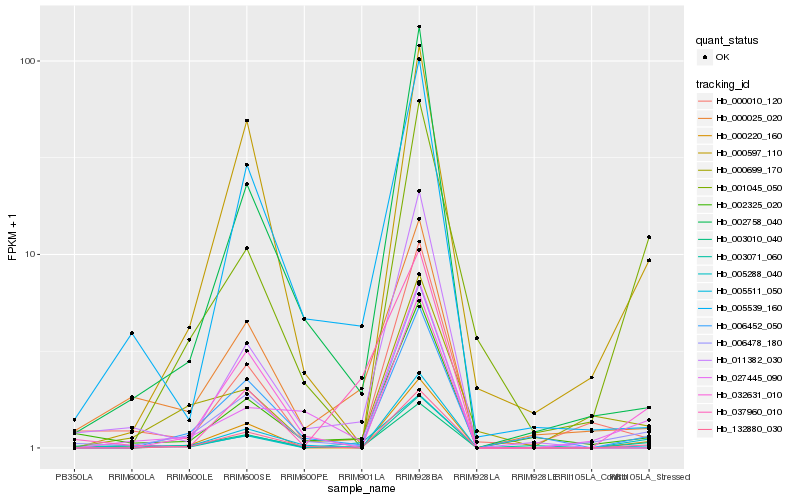
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_040 |
0.0 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_003071_060 |
0.2170603165 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Jatropha curcas] |
| 3 |
Hb_000025_020 |
0.2183050737 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 4 |
Hb_000220_160 |
0.2221394795 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 5 |
Hb_006478_180 |
0.2279612065 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 6 |
Hb_132880_030 |
0.2454952705 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 7 |
Hb_002325_020 |
0.2473190449 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_027445_090 |
0.2479249706 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 9 |
Hb_001045_050 |
0.2556292452 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 10 |
Hb_032631_010 |
0.2594421102 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 11 |
Hb_005539_160 |
0.2653379342 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000597_110 |
0.2655067348 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 13 |
Hb_006452_050 |
0.2657131664 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 14 |
Hb_000010_120 |
0.2686005733 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 15 |
Hb_005511_050 |
0.2699790664 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 16 |
Hb_000699_170 |
0.2712072876 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 17 |
Hb_011382_030 |
0.2725000121 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 18 |
Hb_002758_040 |
0.2727227015 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_003010_040 |
0.2729440536 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 20 |
Hb_037960_010 |
0.274874721 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |