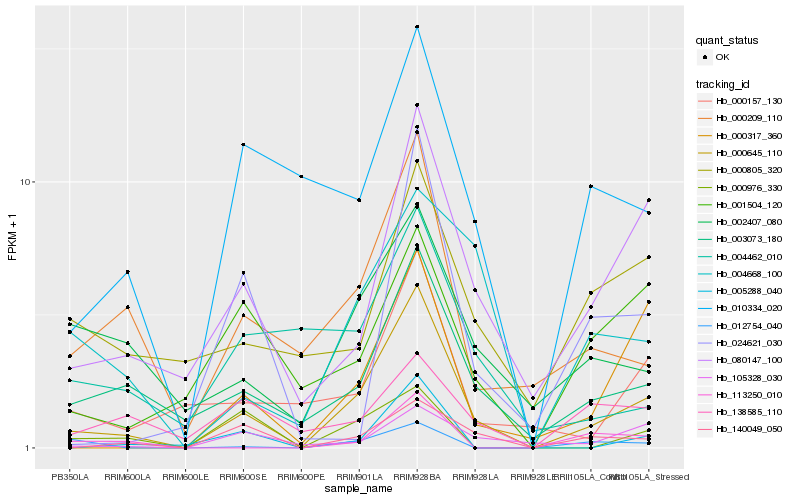
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000645_110 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 2 |
Hb_003073_180 |
0.2223938172 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_080147_100 |
0.2305973428 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 4 |
Hb_140049_050 |
0.2552045226 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 5 |
Hb_000209_110 |
0.2576019316 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 6 |
Hb_000317_360 |
0.2590370765 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g08460 [Jatropha curcas] |
| 7 |
Hb_000157_130 |
0.2769037653 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_105328_030 |
0.2821221414 |
- |
- |
Putative retroelement [Oryza sativa Japonica Group] |
| 9 |
Hb_002407_080 |
0.2830670768 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 10 |
Hb_004668_100 |
0.2833280271 |
- |
- |
- |
| 11 |
Hb_010334_020 |
0.2871800319 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 12 |
Hb_005288_040 |
0.2970313068 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_001504_120 |
0.2975638707 |
- |
- |
- |
| 14 |
Hb_000805_320 |
0.3026059532 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 15 |
Hb_024621_030 |
0.3051931961 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 16 |
Hb_138585_110 |
0.3073958972 |
- |
- |
- |
| 17 |
Hb_113250_010 |
0.3105670391 |
- |
- |
- |
| 18 |
Hb_012754_040 |
0.3110819309 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 19 |
Hb_004462_010 |
0.313957212 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 20 |
Hb_000976_330 |
0.3140055192 |
- |
- |
hypothetical protein POPTR_0001s05180g [Populus trichocarpa] |