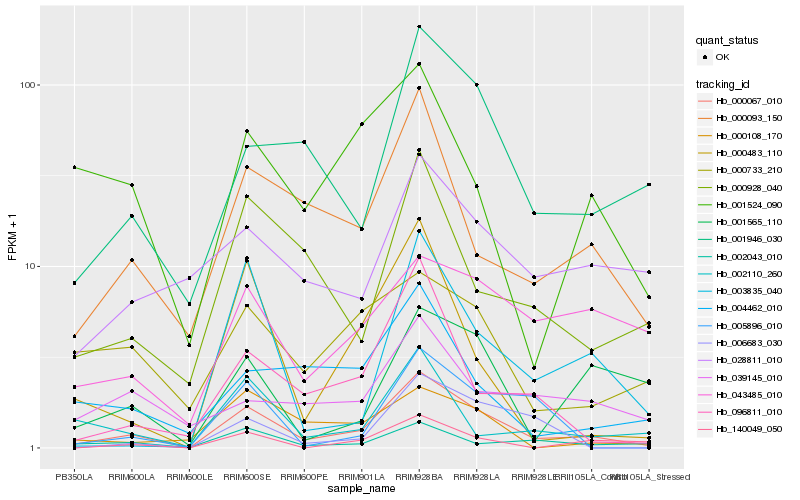
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000067_010 |
0.0 |
- |
- |
hypothetical protein B456_002G147500 [Gossypium raimondii] |
| 2 |
Hb_000093_150 |
0.2203414098 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 3 |
Hb_001946_030 |
0.2246902589 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 4 |
Hb_096811_010 |
0.2420095673 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 5 |
Hb_000928_040 |
0.2474617241 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_002043_010 |
0.2552584519 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_140049_050 |
0.2563154119 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 8 |
Hb_000483_110 |
0.2591184158 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 2, partial [Theobroma cacao] |
| 9 |
Hb_003835_040 |
0.259135985 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 10 |
Hb_004462_010 |
0.2617841011 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 11 |
Hb_000733_210 |
0.2653352489 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 12 |
Hb_002110_260 |
0.2653601252 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_001565_110 |
0.2656697514 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 14 |
Hb_028811_010 |
0.2672549427 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 15 |
Hb_043485_010 |
0.2674048829 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_039145_010 |
0.267851662 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 17 |
Hb_005896_010 |
0.2680756313 |
- |
- |
- |
| 18 |
Hb_000108_170 |
0.2682922757 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 19 |
Hb_006683_030 |
0.2686083159 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 20 |
Hb_001524_090 |
0.2719680057 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |