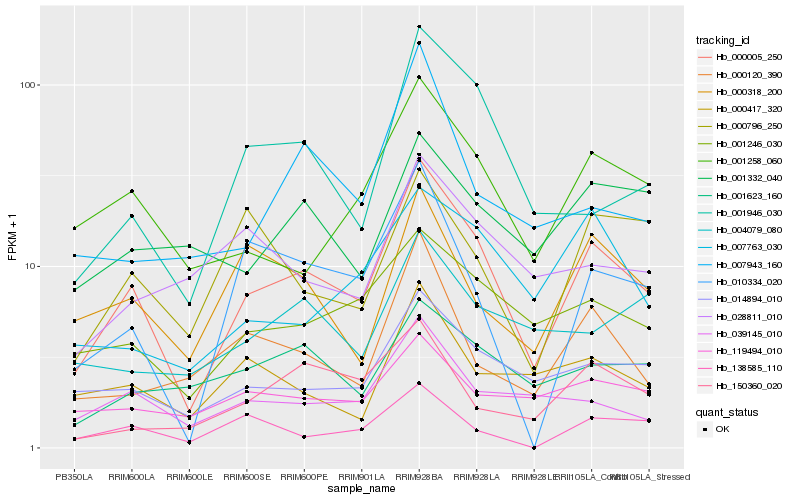
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_014894_010 |
0.17666852 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 3 |
Hb_001258_060 |
0.1769928228 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 4 |
Hb_001246_030 |
0.1803124606 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 5 |
Hb_001946_030 |
0.1866443264 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 6 |
Hb_010334_020 |
0.1951547449 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 7 |
Hb_039145_010 |
0.1976094626 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 8 |
Hb_000120_390 |
0.2014769132 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 9 |
Hb_000318_200 |
0.2046191319 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 10 |
Hb_001623_160 |
0.2056911821 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 11 |
Hb_007763_030 |
0.2080123244 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 12 |
Hb_000417_320 |
0.2097790304 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 13 |
Hb_150360_020 |
0.2145256668 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 14 |
Hb_007943_160 |
0.2160675028 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_138585_110 |
0.2169629021 |
- |
- |
- |
| 16 |
Hb_119494_010 |
0.2170105875 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 17 |
Hb_001332_040 |
0.2172872344 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 18 |
Hb_028811_010 |
0.2175499636 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000796_250 |
0.2204977774 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 20 |
Hb_004079_080 |
0.2210069645 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |