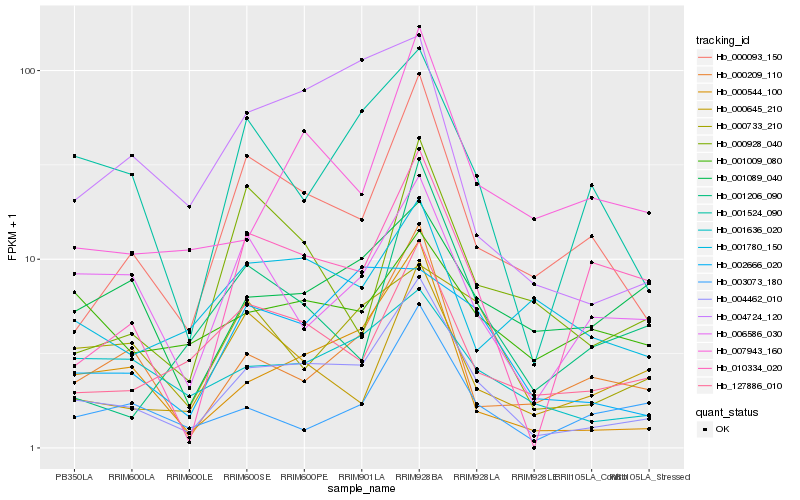
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004462_010 |
0.0 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 2 |
Hb_000544_100 |
0.1679695824 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 3 |
Hb_000093_150 |
0.1761362433 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 4 |
Hb_127886_010 |
0.1866875276 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 5 |
Hb_001524_090 |
0.1953303468 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 6 |
Hb_001636_020 |
0.2010463503 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 7 |
Hb_000209_110 |
0.2044662747 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 8 |
Hb_007943_160 |
0.2087339161 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 9 |
Hb_006586_030 |
0.2112458202 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 10 |
Hb_010334_020 |
0.2170147649 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 11 |
Hb_000645_210 |
0.2173883278 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_004724_120 |
0.2176107958 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 13 |
Hb_003073_180 |
0.2221763667 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_000733_210 |
0.2237366281 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 15 |
Hb_000928_040 |
0.228421168 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002666_020 |
0.2286398794 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 17 |
Hb_001206_090 |
0.2305379507 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 18 |
Hb_001780_150 |
0.2306574173 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 19 |
Hb_001089_040 |
0.2358048616 |
transcription factor |
TF Family: bHLH |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 20 |
Hb_001009_080 |
0.2373355939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |