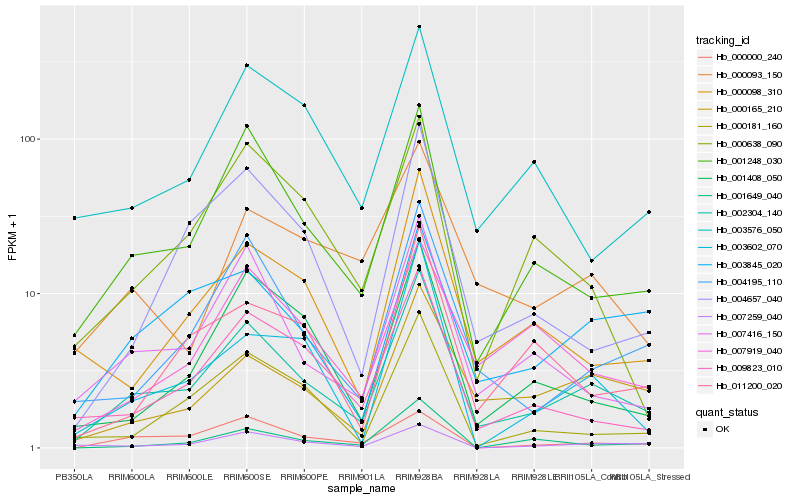
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_140 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_001248_030 |
0.1395831445 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_007416_150 |
0.1521439226 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 4 |
Hb_004195_110 |
0.155043609 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 5 |
Hb_009823_010 |
0.1621294077 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 6 |
Hb_000165_210 |
0.1681277542 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000098_310 |
0.1727722864 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 8 |
Hb_004657_040 |
0.17710364 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 9 |
Hb_001408_050 |
0.1781750633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000181_160 |
0.1795314448 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007259_040 |
0.1824794715 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 12 |
Hb_003602_070 |
0.184217771 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 13 |
Hb_011200_020 |
0.1857909693 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 14 |
Hb_003845_020 |
0.1868421389 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_001649_040 |
0.1880065727 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_007919_040 |
0.1881898689 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 17 |
Hb_003576_050 |
0.1890851898 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 18 |
Hb_000638_090 |
0.1914392233 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000000_240 |
0.1919806101 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 20 |
Hb_000093_150 |
0.1931346078 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |