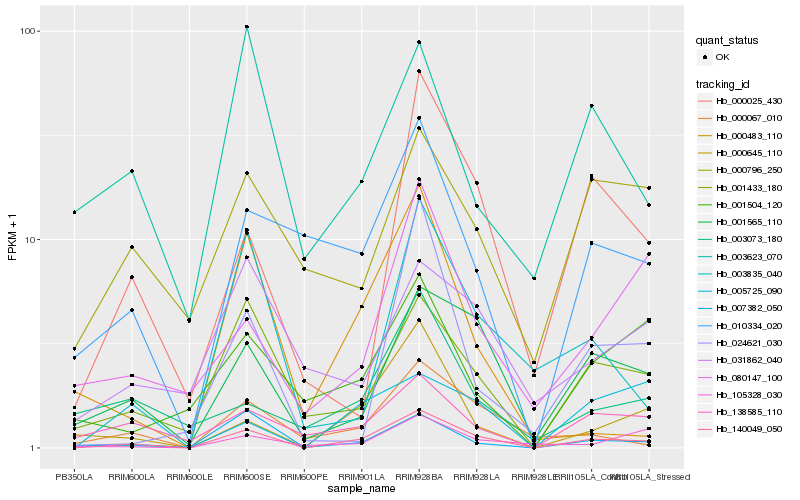
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140049_050 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 2 |
Hb_001565_110 |
0.2229570969 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 3 |
Hb_001433_180 |
0.2361562207 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 4 |
Hb_007382_050 |
0.2480214614 |
- |
- |
PREDICTED: uncharacterized protein LOC105645009 [Jatropha curcas] |
| 5 |
Hb_000645_110 |
0.2552045226 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 6 |
Hb_000067_010 |
0.2563154119 |
- |
- |
hypothetical protein B456_002G147500 [Gossypium raimondii] |
| 7 |
Hb_010334_020 |
0.2612991908 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 8 |
Hb_138585_110 |
0.2662334711 |
- |
- |
- |
| 9 |
Hb_000025_430 |
0.2710221497 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_080147_100 |
0.2931026793 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 11 |
Hb_003835_040 |
0.2944992633 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 12 |
Hb_024621_030 |
0.2962589605 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 13 |
Hb_003073_180 |
0.2964185959 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_001504_120 |
0.2987334011 |
- |
- |
- |
| 15 |
Hb_105328_030 |
0.2999868816 |
- |
- |
Putative retroelement [Oryza sativa Japonica Group] |
| 16 |
Hb_005725_090 |
0.3019202341 |
- |
- |
PREDICTED: phytosulfokines 3 [Cicer arietinum] |
| 17 |
Hb_000796_250 |
0.3055334713 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 18 |
Hb_003623_070 |
0.3059814073 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_031862_040 |
0.3079214044 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 20 |
Hb_000483_110 |
0.3081427974 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 2, partial [Theobroma cacao] |