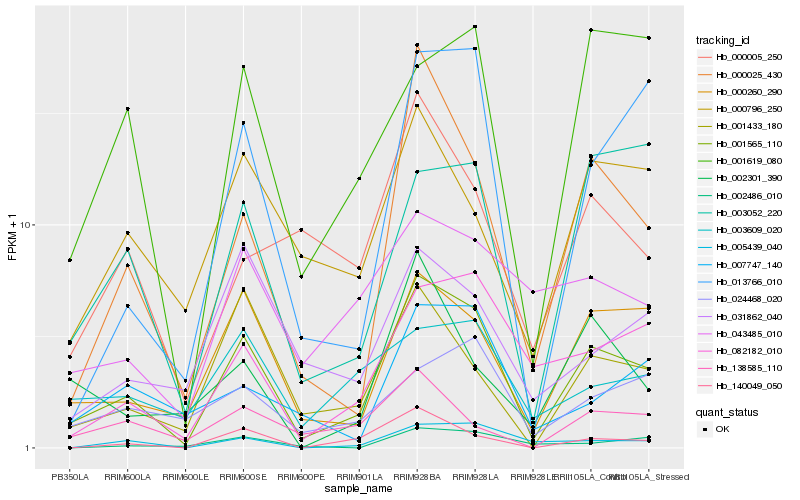
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001565_110 |
0.0 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 2 |
Hb_000260_290 |
0.1837588335 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000025_430 |
0.1975039309 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_013766_010 |
0.2125627302 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 5 |
Hb_082182_010 |
0.2154534984 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_005439_040 |
0.2228246771 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 7 |
Hb_140049_050 |
0.2229570969 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 8 |
Hb_001433_180 |
0.2244761328 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 9 |
Hb_000796_250 |
0.2325488997 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 10 |
Hb_003052_220 |
0.2336441292 |
- |
- |
- |
| 11 |
Hb_001619_080 |
0.2362642425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007747_140 |
0.2394238644 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_003609_020 |
0.2401104849 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 14 |
Hb_024468_020 |
0.2419379983 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 15 |
Hb_000005_250 |
0.242005282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_043485_010 |
0.2421029059 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_138585_110 |
0.2460047068 |
- |
- |
- |
| 18 |
Hb_031862_040 |
0.2477880254 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 19 |
Hb_002301_390 |
0.2522533241 |
- |
- |
- |
| 20 |
Hb_002486_010 |
0.2537678127 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |