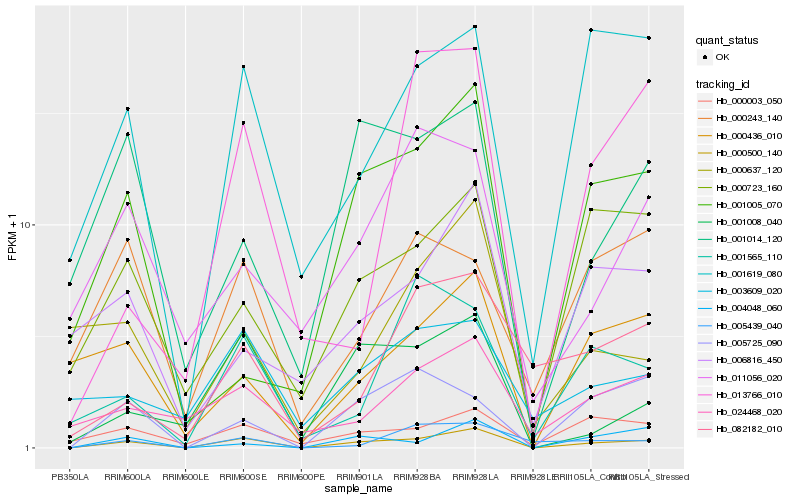
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000500_140 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 2 |
Hb_004048_060 |
0.2367199744 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 3 |
Hb_001619_080 |
0.2453899838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000003_050 |
0.2511761698 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 5 |
Hb_000723_160 |
0.254398265 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 6 |
Hb_001008_040 |
0.2613806772 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 7 |
Hb_000436_010 |
0.264505388 |
- |
- |
PREDICTED: probable polyamine transporter At1g31830 isoform X1 [Eucalyptus grandis] |
| 8 |
Hb_005439_040 |
0.2651117071 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 9 |
Hb_005725_090 |
0.2654006651 |
- |
- |
PREDICTED: phytosulfokines 3 [Cicer arietinum] |
| 10 |
Hb_001014_120 |
0.2655409531 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 11 |
Hb_024468_020 |
0.2693509122 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 12 |
Hb_001005_070 |
0.2726020209 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 13 |
Hb_006816_450 |
0.2760397272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001565_110 |
0.281698922 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 15 |
Hb_003609_020 |
0.2822035593 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 16 |
Hb_082182_010 |
0.2829451218 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_000637_120 |
0.2895574579 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 18 |
Hb_000243_140 |
0.2937577697 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 19 |
Hb_013766_010 |
0.2950679379 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 20 |
Hb_011056_020 |
0.2957575144 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |