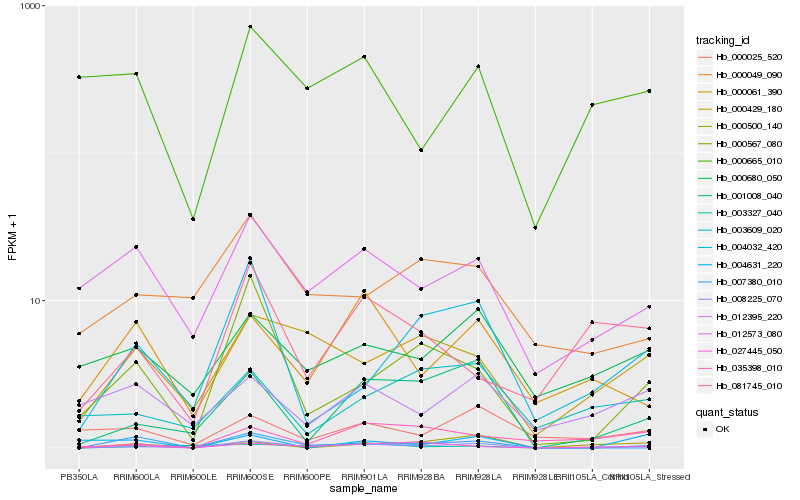
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008225_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_001008_040 |
0.2534437591 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 3 |
Hb_000567_080 |
0.2871544905 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 4 |
Hb_004032_420 |
0.3047746785 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 5 |
Hb_003327_040 |
0.3226069286 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 6 |
Hb_000429_180 |
0.3274841861 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012573_080 |
0.3279632412 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 8 |
Hb_035398_010 |
0.3342124874 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 9 |
Hb_000500_140 |
0.3358070552 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 10 |
Hb_000680_050 |
0.3400041895 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 11 |
Hb_007380_010 |
0.3419335235 |
- |
- |
hypothetical protein EUGRSUZ_L00213, partial [Eucalyptus grandis] |
| 12 |
Hb_000025_520 |
0.346241396 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27110 [Jatropha curcas] |
| 13 |
Hb_027445_050 |
0.3508013498 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 14 |
Hb_000061_390 |
0.3523942773 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 15 |
Hb_004631_220 |
0.3533322952 |
- |
- |
- |
| 16 |
Hb_081745_010 |
0.3533358721 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 17 |
Hb_000665_010 |
0.3573564264 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 18 |
Hb_000049_090 |
0.3575328853 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003609_020 |
0.3581615558 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 20 |
Hb_012395_220 |
0.3593077128 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |