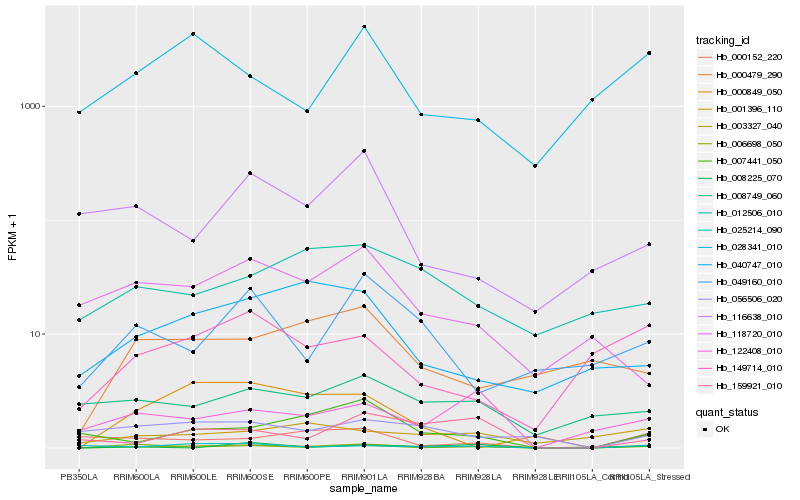
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003327_040 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 2 |
Hb_007441_050 |
0.2734413868 |
- |
- |
unknown [Medicago truncatula] |
| 3 |
Hb_159921_010 |
0.288342248 |
- |
- |
- |
| 4 |
Hb_122408_010 |
0.3004097453 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_040747_010 |
0.3085158704 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 6 |
Hb_008749_060 |
0.3099355252 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 7 |
Hb_149714_010 |
0.3110298628 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 8 |
Hb_000479_290 |
0.3130927096 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X2 [Jatropha curcas] |
| 9 |
Hb_049160_010 |
0.3143837603 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 10 |
Hb_056506_020 |
0.3184404668 |
- |
- |
PREDICTED: uncharacterized protein LOC105647588 [Jatropha curcas] |
| 11 |
Hb_028341_010 |
0.3194722966 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 12 |
Hb_118720_010 |
0.3203173498 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 13 |
Hb_025214_090 |
0.3216383076 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_012506_010 |
0.3224235294 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 15 |
Hb_008225_070 |
0.3226069286 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_000152_220 |
0.3237344919 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 17 |
Hb_000849_050 |
0.3237991846 |
- |
- |
hypothetical protein VITISV_010852 [Vitis vinifera] |
| 18 |
Hb_116638_010 |
0.3272731505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006698_050 |
0.3272930169 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_001396_110 |
0.3295466292 |
- |
- |
DNA primase large subunit, putative [Ricinus communis] |