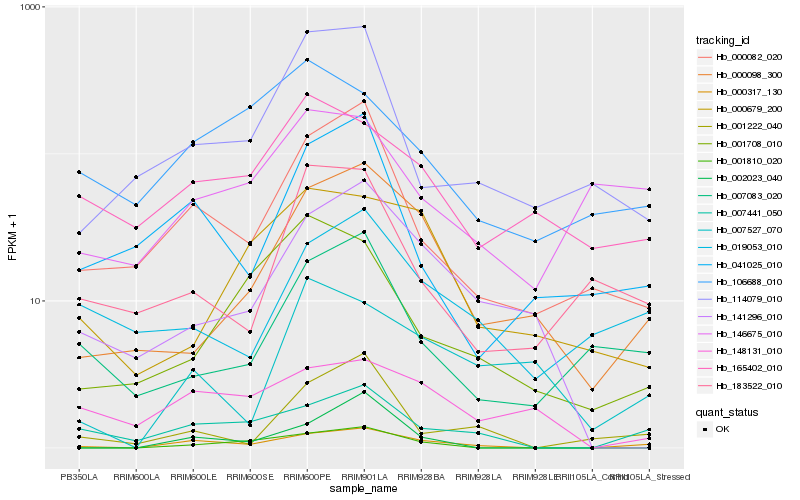
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_130 |
0.0 |
- |
- |
vacuolar membrane protein pep3, putative [Ricinus communis] |
| 2 |
Hb_007441_050 |
0.1903854451 |
- |
- |
unknown [Medicago truncatula] |
| 3 |
Hb_000082_020 |
0.2481074841 |
- |
- |
- |
| 4 |
Hb_000098_300 |
0.2611387464 |
- |
- |
- |
| 5 |
Hb_001222_040 |
0.2690064848 |
- |
- |
- |
| 6 |
Hb_019053_010 |
0.2812928117 |
- |
- |
- |
| 7 |
Hb_001810_020 |
0.2845827281 |
- |
- |
- |
| 8 |
Hb_141296_010 |
0.2846720263 |
- |
- |
- |
| 9 |
Hb_041025_010 |
0.2849813723 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 10 |
Hb_007083_020 |
0.2889561052 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 11 |
Hb_007527_070 |
0.2893999142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 12 |
Hb_146675_010 |
0.2910070324 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 13 |
Hb_148131_010 |
0.2921229978 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12-like, partial [Jatropha curcas] |
| 14 |
Hb_114079_010 |
0.2946385542 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 15 |
Hb_002023_040 |
0.2962411769 |
- |
- |
- |
| 16 |
Hb_000679_200 |
0.3009581746 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 17 |
Hb_106688_010 |
0.3035753996 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 18 |
Hb_165402_010 |
0.3038676989 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 19 |
Hb_183522_010 |
0.3059661608 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 20 |
Hb_001708_010 |
0.3078268381 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |