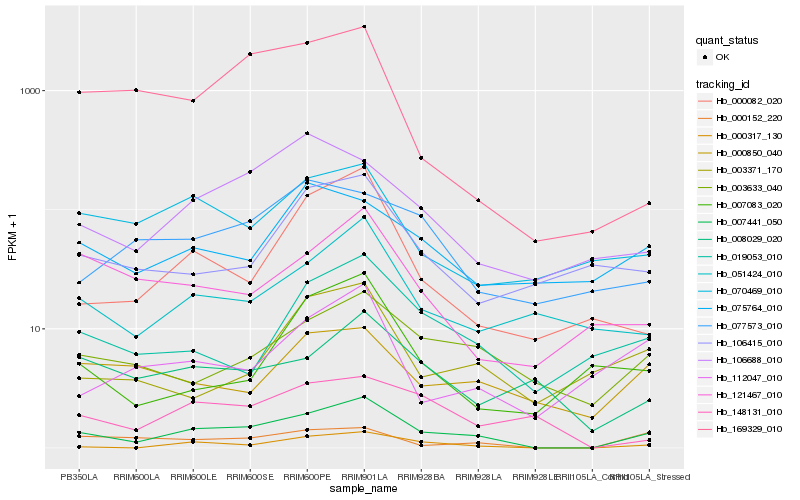
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_050 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_000317_130 |
0.1903854451 |
- |
- |
vacuolar membrane protein pep3, putative [Ricinus communis] |
| 3 |
Hb_003633_040 |
0.2296033033 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 4 |
Hb_019053_010 |
0.2346823243 |
- |
- |
- |
| 5 |
Hb_000152_220 |
0.2348579122 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 6 |
Hb_051424_010 |
0.2388595481 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 7 |
Hb_000082_020 |
0.2458876608 |
- |
- |
- |
| 8 |
Hb_070469_010 |
0.2509792508 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 9 |
Hb_121467_010 |
0.251621216 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 10 |
Hb_106688_010 |
0.2519902129 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 11 |
Hb_008029_020 |
0.253946837 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 12 |
Hb_148131_010 |
0.2539892133 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12-like, partial [Jatropha curcas] |
| 13 |
Hb_112047_010 |
0.2550783902 |
- |
- |
- |
| 14 |
Hb_000850_040 |
0.2576568509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003371_170 |
0.2590451906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_169329_010 |
0.2595887104 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 17 |
Hb_106415_010 |
0.2600195976 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 18 |
Hb_075764_010 |
0.2611419291 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 19 |
Hb_077573_010 |
0.2667518009 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_007083_020 |
0.2669056769 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |