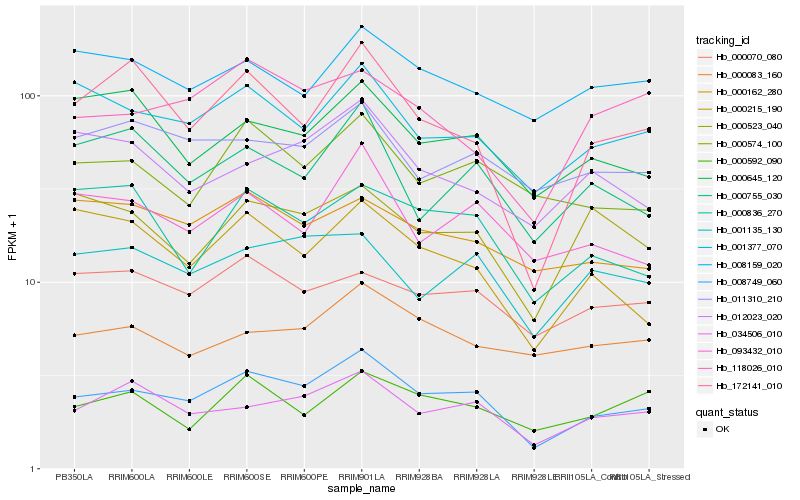
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 2 |
Hb_034506_010 |
0.1029463018 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 3 |
Hb_001377_070 |
0.1078842987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_172141_010 |
0.1157612679 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |
| 5 |
Hb_000574_100 |
0.1187730054 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 6 |
Hb_093432_010 |
0.1203615562 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 7 |
Hb_001135_130 |
0.1205193591 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 8 |
Hb_118026_010 |
0.1263215842 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 9 |
Hb_000215_190 |
0.1277218861 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 10 |
Hb_011310_210 |
0.1277988326 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 11 |
Hb_000070_080 |
0.1294149928 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000523_040 |
0.1296842954 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000755_030 |
0.1302179354 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 14 |
Hb_000162_280 |
0.1304178159 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 15 |
Hb_000645_120 |
0.1308586678 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 16 |
Hb_000836_270 |
0.1314878838 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 17 |
Hb_008159_020 |
0.1315481697 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 18 |
Hb_012023_020 |
0.1316408386 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 19 |
Hb_000592_090 |
0.1325053737 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18020 [Jatropha curcas] |
| 20 |
Hb_000083_160 |
0.1325995736 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |