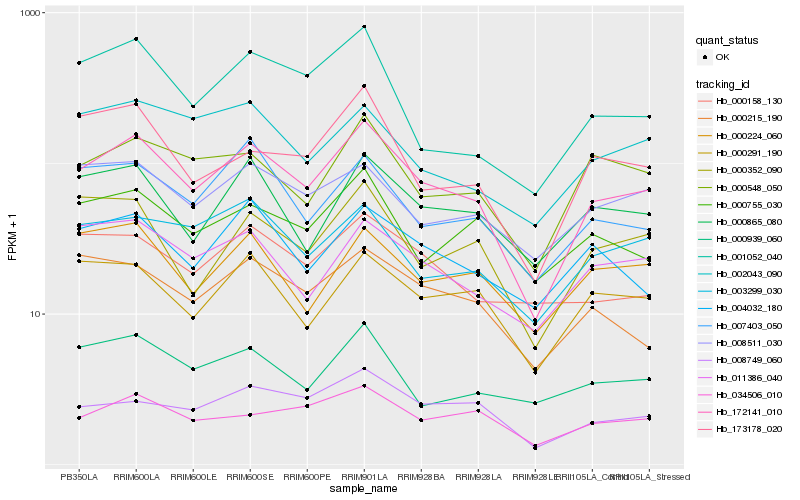
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172141_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |
| 2 |
Hb_008749_060 |
0.1157612679 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 3 |
Hb_011386_040 |
0.1167936536 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000548_050 |
0.1177964727 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 5 |
Hb_000291_190 |
0.1188404931 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 6 |
Hb_000865_080 |
0.1202092676 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 7 |
Hb_007403_050 |
0.1209741351 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 8 |
Hb_003299_030 |
0.1216077978 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 9 |
Hb_173178_020 |
0.1229000704 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000352_090 |
0.1237331182 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001052_040 |
0.1241909342 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000215_190 |
0.1242965249 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 13 |
Hb_004032_180 |
0.1247710579 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 14 |
Hb_008511_030 |
0.1262012044 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 15 |
Hb_002043_090 |
0.1281861596 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 16 |
Hb_000158_130 |
0.1283019037 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000224_060 |
0.1284064892 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 18 |
Hb_000755_030 |
0.1285796278 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 19 |
Hb_034506_010 |
0.1286610237 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 20 |
Hb_000939_060 |
0.1306958815 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |