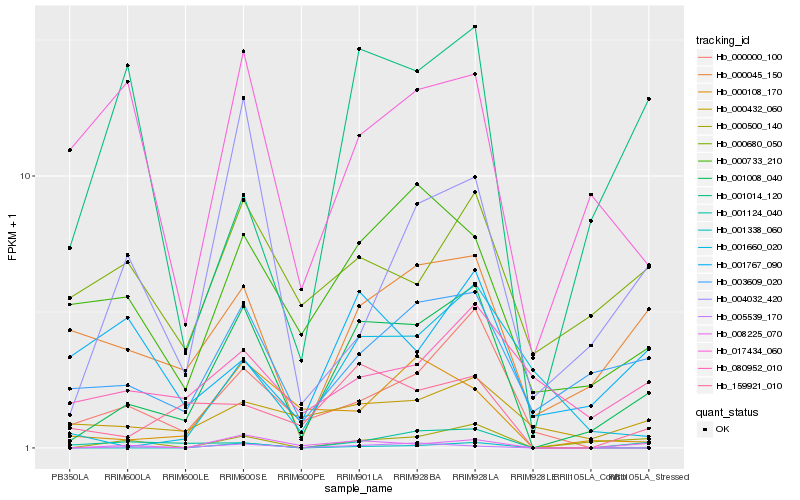
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_040 |
0.0 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 2 |
Hb_003609_020 |
0.2470818663 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 3 |
Hb_008225_070 |
0.2534437591 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_000045_150 |
0.2583917333 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 5 |
Hb_000500_140 |
0.2613806772 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 6 |
Hb_000733_210 |
0.2741282334 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 7 |
Hb_159921_010 |
0.2802810885 |
- |
- |
- |
| 8 |
Hb_004032_420 |
0.2827999325 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 9 |
Hb_001124_040 |
0.2856071115 |
- |
- |
PREDICTED: uncharacterized protein LOC103413595 [Malus domestica] |
| 10 |
Hb_005539_170 |
0.2933608748 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 11 |
Hb_000108_170 |
0.2933943666 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 12 |
Hb_000432_060 |
0.299440919 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000000_100 |
0.3034530642 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 14 |
Hb_001014_120 |
0.3061615425 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 15 |
Hb_017434_060 |
0.3104421651 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 16 |
Hb_001660_020 |
0.3135922465 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 17 |
Hb_080952_010 |
0.3144066808 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 18 |
Hb_001767_090 |
0.3167186994 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_000680_050 |
0.3170595034 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 20 |
Hb_001338_060 |
0.3171772582 |
- |
- |
- |