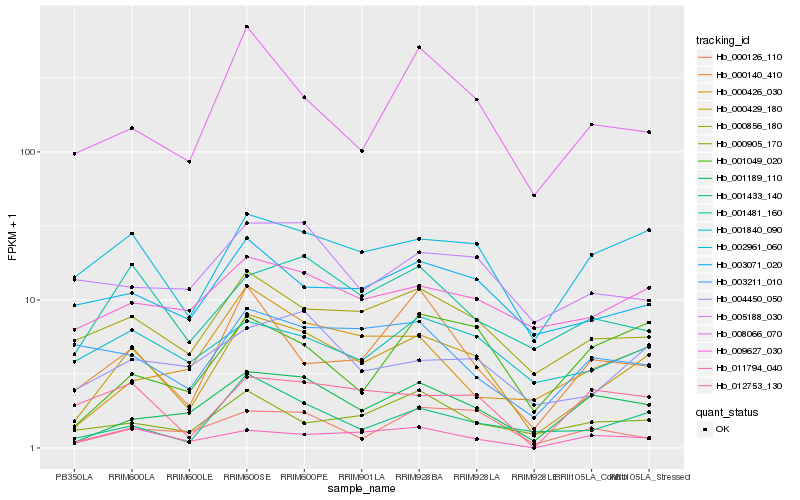
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000429_180 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002961_060 |
0.1566622034 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 5 [Populus euphratica] |
| 3 |
Hb_000856_180 |
0.1660546871 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_001433_140 |
0.1801544457 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 5 |
Hb_001481_160 |
0.1816953286 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 6 |
Hb_001189_110 |
0.1822561505 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 7 |
Hb_004450_050 |
0.186183942 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 8 |
Hb_003071_020 |
0.1909328412 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 9 |
Hb_011794_040 |
0.1922968772 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 10 |
Hb_009627_030 |
0.1940107325 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 11 |
Hb_001840_090 |
0.1959052276 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_008066_070 |
0.1970794614 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 13 |
Hb_012753_130 |
0.2024044348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000140_410 |
0.2041699939 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000905_170 |
0.204873512 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_001049_020 |
0.2056153779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000426_030 |
0.2110945357 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003211_010 |
0.212924024 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 19 |
Hb_005188_030 |
0.2135485722 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000126_110 |
0.2135620813 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |