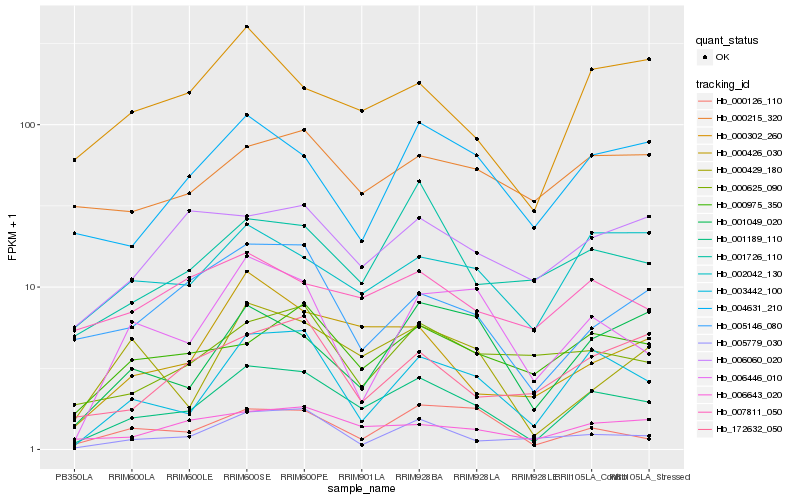
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001189_110 |
0.0 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 2 |
Hb_003442_100 |
0.13796607 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 3 |
Hb_006643_020 |
0.160844339 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 4 |
Hb_000426_030 |
0.1721067303 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000975_350 |
0.1734956648 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_000302_260 |
0.1756108315 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 7 |
Hb_002042_130 |
0.179578634 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 8 |
Hb_004631_210 |
0.1807154027 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 9 |
Hb_001049_020 |
0.1809363279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000429_180 |
0.1822561505 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001726_110 |
0.1822903118 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 12 |
Hb_005146_080 |
0.1840523608 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005779_030 |
0.1842914951 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 14 |
Hb_000126_110 |
0.1850215771 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 15 |
Hb_006060_020 |
0.1851165533 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000625_090 |
0.1870923815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006446_010 |
0.1882967711 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_172632_050 |
0.1916084937 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 19 |
Hb_007811_050 |
0.1941518724 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 20 |
Hb_000215_320 |
0.194178248 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |